Abstract
There is growing evidence that regions of the genome that cannot encode proteins play an important role in diseases. These regions are usually transcribed into long non-coding RNAs (lncRNAs). LncRNAs, little or no coding potential, are defined as capped transcripts longer than 200 nucleotides. New sequencing technologies have shown that a large number of aberrantly expressed lncRNAs are associated with multiple cancer types and indicated they have emerged as an important class of pervasive genes during the development and progression of cancer. However, the underlying mechanism in cancer is still unknown. Therefore, it is necessary to elucidate the lncRNA function. Notably, many lncRNAs dysregulation are associated with Oral squamous cell carcinoma (OSCC) and affect various aspects of cellular homeostasis, including proliferation, survival, migration or genomic stability. This review expounds the up- or down-regulation of lncRNAs in OSCC and the molecular mechanisms by which lncRNAs perform their function in the malignant cell. Finally, the potential of lncRNAs as non-invasive biomarkers for OSCC diagnosis are also described. LncRNAs hold promise as prospective novel therapeutic targets, but more research is needed to gain a better understanding of their biologic function.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC), characterized by differentiation and a propensity for lymph node metastasis [1], is the sixth most common cancer worldwide with over 200,000 newly diagnosed once each year, and can be divided into three major subsites: buccal mucosal SCC (BMSCC), tongue SCC (TSCC), and lip SCC (LSCC) [2, 3]. Percentages of morbidity and mortality in males are 6.6/100,000 and 3.1/100,000 respectively, while in females, the same percentages are 2.9/100,000 and 1.4/100,000 [4]. Additionally, the incidence of OSCC is increasing among young white individuals age 18 to 44 years, particularly among white women [5]. Due to its risk factor exposure, low cure rate and high mortality, OSCC represents a global public health problem, with a great individual and socioeconomic burden.
The occurrence of OSCC is a complex multistep process. Normal oral keratinocytes are prolonged by adverse factors, resulting in intracellular microenvironment imbalance and genetic alterations. Genetically unstable precancerous keratinocytes can transfer these inheritable alterations to their clones [6]. These carcinogenic factors include key disorders on TP53, NOTCH1 (Notch homolog 1 genes are translocation-associated), EGFR (epidermal growth factor receptor), CDKN2A (cyclin-dependent kinase inhibitor 2a), STAT3 (signal transducer and activator of transcription 3), Cyclin D1 and retinoblastoma [7]. Normal oral keratinocytes are transformed into precancerous lesions through various signaling pathways, which further deteriorate into malignant tumors. Moreover, Yuan et al. [8] performed a case-control study including 444 OSCC cases and 984 healthy controls to investigate whether H19 genetic variants affect the risk of OSCC in the Chinese population. It was finally determined that the two SNPs, rs2839701 and rs217727, were related to OSCC susceptibility and indicated that the SNPs in H19 might be OSCC biomarkers.
Smoking and drinking are the two main causes of the high incidence of OSCC. A meta-analysis showed that smokers were at higher risk of developing oral cancers than non-smokers [9]. Cigarette smoke exerts inflammatory and suppressive effects on immune cells, alters mucosal immunity and promotes autoimmunity, resulting in oral cancers [10]. Yamashita et al. [11] found that smoking and drinking can inhibit 5-fluorouracil (5-FU)-related metabolic enzymes through the induction of dihydropyrimidine dehydrogenase (DPD; a sole catabolic enzyme of 5-FU) activity, which in turn lead to oral cancers. In addition, alcohol consumption can interact with the polymorphisms of ALDH2 and CYP2E1-RsaI genes to increase OSCC risk [12]. Human papillomavirus (HPV) is also considered as one of the potential risk factors of OSCC. As early as 2007, HPV 16 was recognized by the International Agency for Research on Cancer as a risk factor for OSCC. HPV33, HPV35 and others (also found in cervical cancer) have been also considered to trigger OSCC [13].
Despite advancements in diagnosis and treatment methods, 5-year survival rate has not improved significantly over the past decade, which ranges from 45 to 50% [14]. Surgical resection is considered to be a promising treatment strategy for early cancer [15]. However, recurrence after surgical resection is still a serious cause of cancer-related death [16]. Therefore, how to prevent postoperative recurrence and improve patient survival is still a major challenge in OSCC treatment. Increasing evidence shows that multiple oncogenes and tumor suppressor genes are involved in OSCC. This contributed to better understand the exact mechanisms between lncRNAs and OSCC, providing suitable approaches for clinical treatment. Meanwhile, the discovery of new cancer molecular targets can also effectively help to understand the pathogenesis and prognosis of OSCC.
Accumulating evidence showed that non-coding RNAs (ncRNAs), such as long non-coding RNAs (lncRNAs), played vital regulatory roles in the cellular physiological process [17,18,19] (Fig. 1). For example, lncRNAs could act as miRNAs sponge to weaken regulations of miRNAs on mRNAs [20, 21]. Regulatory mechanism of lncRNA-induced oral cancers, especially OSCC, should not be ignored. LncRNAs associated with cancer pathogenesis were primarily involved in cellular macromolecules (including chromatin, protein, RNA) [22, 23]. So far, no perfect diagnostic marker of OSCC has appeared. Aberrant expression of some lncRNAs had been shown to be closely correlated with cancer prognosis. In OSCC tissues, HOTAIR (HOX transcription antisense RNA) was highly expressed, and its expression level was correlated with tumor size, TNM (Tumor Node Metastasis) stage, and prognosis of OSCC [24]. This suggested that HOTAIR could be employed as a biomarker for diagnosis and prognostic determination as well as a molecular target for therapy. On the one hand, epigenetic changes in a body (mainly involved in DNA methylation, histone modifications and modifications in micro ribonucleic acids) can provide valuable biomarkers [25]. On the other hand, comparing seven mRNAs and three proteins in saliva, OSCC patients had a higher level of interleukin (IL)-8 and subcutaneous adipose tissue than healthy controls [26]. This result suggested new biomarkers. Moreover, Tang et al. [27] found that MALAT-1 and HOTAIR in saliva samples from OSCC patients could be expressed in patients with primary tumor. It was worth noting that the expression level of HOTAIR in saliva of OSCC metastatic patient was different from that of primary tumor controls. This suggested that the detection of lncRNAs in saliva can be used for clinical non-invasive and rapid diagnosis of OSCC, and to determine whether there was metastasis. At present, most of the clinical treatment programs (chemoradiotherapy, surgery, EGFR inhibitors and COX-2 inhibitors, and photodynamic therapy) are high economic cost and highly damaging treatment, which are a burden for patients and society [28, 29]. Nanotechnology-Based approaches for prevention and therapy have become a hot spot and need further clinical validation [30]. In general, an effective, safe and prognostic treatment is urgently needed for the therapy of OSCC. Therefore, precise treatment is a must. The role of ncRNAs, particularly lncRNAs, in cancer is gradually being amplified, and the underlying mechanism between them and OSCC needs to be clarified [31]. Therefore, this review aims to elucidate the mechanism of lncRNAs regulation in OSCC patients, and to explore a new approach for better clinical treatment of diseases.
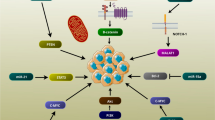
LncRNAs exert functions through a variety of signaling pathways in the human body. a miRNA sponge. MIAT, acting as a molecular sponge, binds to miR-150-5p, thereby upregulating the level of miR-150-5p target gene. b mRNA stability/degradation. LncRNA binding to mRNA may stabilize (e.g., BACE1-AS prevents miRNA-induced repression of BACE1 transcript) or decay target transcripts. c Translation. LncRNAs promote (like antisense Uchl) or repress (like lincRNA-p21) translation of transcripts. d Alternative splicing. MALAT1 acting as scaffold for SR proteins regulates pre-mRNA alternative splicing. e Transcription. PACER (lethe and p50-associated Cox-2 extragenic RNA) directly interacts with different subunits of NF-κB, thus preventing it from binding to the Cox-2 promoter. THRIL, together with heterogeneous nuclear ribonucleoproteins (hnRNPs), acts as RNA-protein complex and binds to TNF-α promoter and induces TNF-α expression. f Epigenetic imprinting. Working models of gene regulation by cis- (a) and trans-acting (b) lncRNAs. LncRNAs, such as Xist/RepA, Air, HOTAIR, and Kcnq1ot1, may act as docking platforms for the chromatin remodeling complex, polycomb repressive complex (PRC2) 2, which methylates histone H3 at lysine 27 (H3K27me3), leading to a repression or gain of transcriptional activity, respectively
Overview of lncRNA
LncRNAs are a novel class of ncRNAs and pervasively transcribed in the human genome. Most (but not all) lncRNAs are synthesized by RNA polymerase II and share many of the biological characteristics of mRNAs, though they bear little or no coding potential [32]. Unlike siRNAs and miRNAs whose sizes are usually comprised between 20 to 24 nucleotides, lncRNAs range in size from 200 to more than one hundred thousand nucleotides and are always capped and polyadenylated [33, 34].
The large number of lncRNAs, large molecular weight and poor stability in vitro have hindered the revealing of its structure. The nucleotide sequence of lncRNAs constitutes its primary structure. LncRNAs can regulate transcriptional translation of a target gene directly or a gene upstream or downstream of a target gene indirectly by binding to a target gene by base-complementary pairing [35], like lncRNA Gas5 [36] and lncRNA 1/2-sbsRNAs [37]. Meanwhile, a major feature of lncRNAs is a propensity to fold into thermodynamically stable secondary and higher-order structures. Hydrogen bonds formed by internal structure of RNA (including the Watson-Crick face, the Hoogsteen and ribose face) together construct the secondary its structure that include double helices, hairpin loops, bulges and pseudoknots [38, 39] (Fig. 2). The secondary and higher-order structures of RNA appear to play their primary biological function. For example, p53 is activate by motifs M2 and M3 that are secondary folding motifs of lncRNA MEG3 (maternally expressed gene 3) isoforms, rather than its primary sequence [40].
The steroid receptor RNA activator (SRA) lncRNA contains 4 subdomains. The human SRA has a length of 870 nt, organized into 4 sub-domains and 25 helices. Four biochemical techniques (SHAPE, in-line, DMS and RNase V1) were used to obtain the secondary structure. Blue, domain I; green, domain II; black, domain III; yellow, domain IV
By using antisense oligonucleotides and RNA interference, it has been shown that lncRANs are present in different parts of the cell, including the nucleus (such as: MALAT1 and NEAT1), cytoplasm (such as: DANCR and OIP5-AS1) or both (such as: TUG1, CasC7 and HOTAIR) [41]. Technological advances have enabled us to explore the vital roles of the lncRNA family (Table 1) [42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63]. Specifically, they were mainly divided into in vivo (DMS-seq, Structural-seq, and Mod-seq, icSHAPE, CLASH and hiCLIP) and in vitro (SHAPE-seq, SHAPE-MAP, and RING-Map, PARS and FragSeq, ss/dsRNA-seq Techniques) methods [64]. These techniques, especially combined with high-throughput sequencing, were also speculated to investigate lncRNA multi-level structures [65, 66].
Accompanying with the increasing number of lncRNAs, they can be mainly divided into the following categories according to different characteristics: (1) genome location and context (intergenic lncRNAs and intronic lncRNAs, sense and antisense lncRNAs), (2) exerted an effect on DNA sequences (cis-lncRNAs, trans-lncRNAs), (3) mechanism of functioning (transcriptional regulation, post-transcriptional regulation and other mechanisms of lncRNA functioning), (4) targeting mechanism [67, 68].
To date, it has demonstrated lncRNAs could regulate multiple disease progressions. For instance, 4313 lncRNAs were upregulated and 4612 lncRNAs were downregulated in periodontitis by using RT-PCR [69]. Additionally, the upregulation of lncRNA SNHG20 and the downregulation of DLEU1 (deleted in lymphocytic leukemia 1) were stably correlated with the progression of OSCC. Nishiyama et al. [70] found DLEU1 silencing suppressed migration, invasion, and xenograft formation in OSCC cells, which was suggestive of its oncogenic functionality. Another lncRNA, UCA1 (urothelial cancer associated 1), also was upregulated in OSCC and enhanced proliferation and metastasis of OSCC cells [71], which was similar to consequences of other cancers in lung [72], stomach [73] and bladder [74].
LncRNA-centric targeting regulation
LncRNAs can mediate chromatin remodeling and transcription regulation, mainly as signals, decoys, guides, and scaffolds, resulting in the downregulation or upregulation of target genes and triggering various diseases [75, 76] (Fig. 3). For example, lncRNA can interact with DNA. The locus 515 kb upstream of MYC can transcribe CCAT1-L, which can interact with MYC transcriptional regulation and accelerate long-range chromatin looping. Specifically, in cis overexpression of CCAT1-L triggers tumorigenesis through promoting MYC expression [77]. In addition, lncRNA-RNA interactions and lncRNA-protein interactions together constitute a complex regulatory network of lncRNA to control occurrence and development of cancers [78, 79]. Xu et al. [80] elaborated that various lncRNAs are involved in the pathogenesis of prostate cancer and can be used as biomarkers for diagnosis, treatment and prognosis. This also suggested that lncRNAs can play a similar role in OSCC.
LncRNAs dysregulation is associated with a variety of diseases in humans. LncRNAs can affect human health through a variety of signaling pathways which can be divided into cytoplasmic signaling pathways and intranuclear signaling pathways. Cytoplasmic signaling pathways include mRNA stability, miRNA sponge and mRNA translation. Intranuclear signaling pathways include splicing, histone modification, chromatin interaction and transcriptional regulator
“Competing endogenous RNA (ceRNA)” have been proposed to emphasize regulatory dialogues between different RNAs, including lncRNAs, miRNAs, transcribed pseudogenes, and circular RNAs (circRNAs) [81]. Particularly, the focus of interactions between miRNAs and lncRNAs in various human disease progression is gaining attention. MiRNAs, composed of 19–25 base pairs, mainly target protein-coding genes at the post-transcriptional level [82]. For example, Hsa-miR-1 suppresses expression of the UCA1 via an Ago2-slicer-dependent signaling and structure recognition 3-untranslated regions (3-UTRs) of UCA1 to play tumor suppressive roles [83]. Similar and different mechanisms also occur in pancreatic cancer [84], breast cancer [85] and colorectal cancer [86]. Generally speaking, lncRNAs exert “sponge-like” effects on various miRNAs to inhibit miRNA-mediated functions (Fig. 4). However, regulatory networks of lncRNAs still have unknown areas.
Biologic functions of lncRNAs in OSCC
The carcinogenesis of OSCC is a multifactorial and multistep process, involving various aspects such as genes, epigenetics and environment. With the continuous exploration and clarification of the structure and function of lncRNAs, the dysregulation of lncRNAs has become a non-negligible regulatory element for the development of cancer [87]. Hereby, this review will elaborate on the regulation of lncRNAs in OSCC (Table 2) [70, 88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106].
Molecular mechanisms of lncRNAs in OSCC
MALAT1, mapped to human chromosome 11q13 and 8.7 kb long, was a highly conserved lncRNA and was also referred to as NEAT2 [107,108,109]. The triple helix-structure at the 3’end of MALAT1 given it unique stability [110, 111]. MALAT1 might interact with SR (serine/arginine-rich) splicing factors (including SRSF1, 2, and 3), which were involved in exon recognition and alternative splicing, to regulate alternative splicing of a subset of pre-mRNAs [112, 113]. MALAT1 was also involved in transcriptional and post-transcriptional regulation [114]. Moreover, binding of methylated and unmethylated Polycomb 2 protein to MALAT1 controlled relocation of growth control genes between Polycomb bodies and interchromatin granules [115]. It had been observed that level of MALAT1 was aberrant in some human tumors [107, 114, 116], and its expression level was associated with tumor recurrence and metastasis. Studies had shown that MALAT1 promoted cellular proliferation by regulating the activity of the E2F1 transcription factor, and then enhanced tumorigenesis [117, 118]. E2F1 transcription factor affected cell cycle regulation and apoptosis [119]. Meanwhile, depletion of MALAT1 affected activity of the oncogenic transcription factor B-MYB (MYBL2), blocking cell cycle in G1/S phase, thereby reducing cell proliferation [118, 120]. B-MYB was a physiological regulator of cell cycle progression, cell survival and cell differentiation, and its overexpression was associated with poor patient outcome in numerous cancers [121]. These studies indicated that MALAT1 played important functions in a string of biological processes.
Emerging evidence suggested that epithelial–mesenchymal transition (EMT), an indispensable mechanism during morphogenesis, was also a crucial event in OSCC. After SDF-1/CXCR4 system induction, EMT may activate PI3K-AKT/PKB signaling pathway to participate in the lymph node metastasis of OSCC [122]. When MALAT1 was deleted in OSCC cell lines TSCCA and Tca8113, EMT mediated cell migration and invasion were inhibited. The low level of MALAT1 weakened β-catenin and NF-κB signaling pathways in OSCC. Correspondingly, tumor growth in the Tscca xenograft model was also inhibited [88]. In addition to MALAT1 increased, STAT3 was also overexpressed, while miR-125b was downregulated in OSCC cell lines [123]. STAT3 was a molecule in the OSCC inflammation-mediated/related carcinogenesis signaling pathways. It can regulate the expression of various genes to cope with cellular stimuli. Meanwhile, STAT3 and NF-κB interacted with each other to regulate cell tumor angiogenesis and invasiveness [124]. Furthermore, miR-125b can directly bind to the 3′-UTR of STAT3, and then decreased the protein levels of STAT3 in MG-63 and Saos-2 cells, suggesting that STAT3, as the functional downstream target of miR-125b, played a role in the transcriptional activation of miR-125b [125]. MALAT1 attenuated the tumor suppressive effect of miR-125b mimics by up-regulating STAT3. The established nude mouse model was further confirmed that upregulated MALAT1 played an oncogene role in OSCC via miR-125b/STAT3 axis [89]. These newly discoveries need further investigation to exploit mechanistic insights of MALAT1 in OSCC progress.
Colon Cancer Associated Transcript 1 (CCAT1), also known as a cancer-associated region lncRNA-5 (CARLo-5) or CCAT1-S with a length of 2628 nucleotides, located in chromosome 8q24.21 [77]. CCAT1 contained two exons and a poly-A tail and was mainly expressed in the nucleus. On the one hand, the CCAT1-L locus is located within a strong super-enhancer that consist of large clusters of transcriptional enhancers formed by binding of master transcription factors/mediators and to be associated with genes that control and define cell identity [77]. On the other hand, CCAT1 closed to c-Myc, a well-known oncogenic transcription factor, and was triggered by c-Myc, resulting in cell proliferation and invasion enhancing [126]. C-Myc, one of the Myc proto-oncogene family members, was found a positive correlation with STAT3 [127]. CCAT1 could trigger c-Myc overexpression through its ceRNA activity on miR-155 [128]. It was found that the high level of CCAT1 downregulated miR155-5p, let7b-5p, miR490-3p by a sponging mechanism and miR218-5p by epigenetic silencing [21, 129]. CCAT1 had been shown to be overexpressed in a variety of cancers and a rising star of oncogenic lncRNAs [128, 130, 131].
Arunkumar et al. [90] collected 60 OSCC tumor samples and eight normal tissue samples, and found that c-Myc was also overexpressed in CCAT1 overexpressing tumor tissues, while miR155-5p and let7b-5p were downregulated. MiR218-5p and miR490-3p were also low expressed due to CCAT1 acted as a sponge. Moreover, miR-155-5p inhibitor, as an EMT suppressor, suppressed the STAT3 signaling pathway and increased suppressor of cytokine signaling 1 (SOCS1) expression. Suppressor of cytokine signaling 1 (SOCS1) recently served as a novel miR-155 target in breast cancer, and might also exerted roles in OSCC [132]. Geng et al. [91] explored the potential effects of Porphyromonas gingivalis (P. gingivalis) on OSCC and found that long-term stimulation of P. gingivalis promoted cell proliferation, accelerated cell cycle and promoted cell migration and invasion abilities. Further, CCAT1 was upregulated by using validation of microarray and proteomic assay. Long-term exposure of P. gingivalis can trigger tumor-associated molecules, such as CCAT1, to enhance tumorigenic properties of human immortalized oral epithelial cells (HIOECs) and participate in the pathogenesis of OSCC [91].
MEG3, the first to be found to have tumor suppressive effects, was a maternally imprinted gene located on chromosome 14q32.3 within DLK1–MEG3 locus [133, 134]. It was also called gene trap locus2 (Gtl2) located at chromosome in mouse [135]. The MEG3 gene, was controlled by two differentially methylated regions (DMRs) that comprised of multiple methylated CpG sites: the intergenic DMR (IG-DMR) and the MEG3-DMR [136]. Multiple signaling pathways were involved in MEG3 inhibition of cell proliferation and metastasis [137,138,139]. Elevated levels of MEG3 or/and miRNA-26a inhibited cell proliferation, suppressed cell cycle progression and induced cell apoptosis [140].
After treating HOK cells with arecoline, Shiah et al. [92] found a significant decrease in MEG3 and 14q32.2 miRNAs. While Wnt-7b overexpressed, the phosphorylation of GSK-3β and active-β-catenin in DOK cells were markedly enhanced, causing cyclin D and c-Myc upregulated [92]. Cyclin D1 was an indispensable nuclear protein in the G1/S phase of the cell cycle. Bova et al. founded that cyclin D1 overexpression and cyclin D1 gene amplification in OSCC [141]. In OSCC cells, multiple signaling pathways, including DNA methylation, downregulated MEG3 expression. MEG3 decreased in OSCC cells by using RT-qPCR technique. Low expression of MEG3 significantly increased Cal27 cell proliferation when compared with control group, suggesting that MEG3 suppressed OSCC cell proliferation. Meanwhile, OSCC cell apoptosis was inhibited, and metastasis was promoted [93]. Wnt/β-catenin signaling pathway, one of the classical signaling pathways in the process of cell signal transduction, was involved in cancer cell proliferation, migration, invasion, tumorigenesis and metastasis [142, 143]. Taken together, MEG3 was found that it could inhibit the Wnt/β-catenin axis to act as a tumor suppressor [93].
Urothelial carcinoma-associated 1 (UCA1) was located on human chromosome 19p13.12 and contained three exons and two introns. UCA1 had three isoforms, including 1.4 kb, 2.2 kb, and 2.7 kb in length, generated by splicing and polyadenylated [144]. The 1.4 kb isoform was labeled as lncRNA UCA1; the 2.2 kb isoform was labeled lncRNA UCA1a or lncRNA CUDR; while the biological role of the 2.7 kb isoform was not known [145]. Additionally, UCA1 may affect CREB expression and activity through PI3K-AKT dependent pathway, and then regulated cell cycle progression [146]. The PI3K/AKT/mTOR signaling pathway was activated in various cancers via stimulation of proliferation, survival, metabolic reprogramming, and invasion/metastasis, as well as suppression of autophagy and senescence [147, 148]. Ectopic expression of lncRNA UCA1 in bladder cancer cell line BLS-211 demonstrated that UCA1 was oncogenic [149]. Studies had found UCA1 was dysregulated and participated in the development of a few cancers including hepatocellular carcinoma [150], pancreatic cancer [145], bladder cancer [74].
Yang et al. [94] concluded that UCA1 was upregulated from 140 TSCC tissue samples. The results demonstrated that tumor growth was inhibited in vivo after UCA1 was deleted. In addition, UCA1 silencing inhibited cell proliferation, migration and invasion in OSCC cell lines. Correspondingly, UCA1-si could suppress OSCC cell proliferation in vitro via the CCK-8 assay. Further analysis found UCA1 upregulation could activate the Wnt/β-catenin signaling pathway [94]. In another study, the results suggested UCA1 overexpressed in OSCC tissues, cell lines, and Cisplatin (CDDP)-resistant OSCC cells by qRT-PCR [95]. CDDP was an anti-tumor drug that was clinically used to treat OSCC [151]. After UCA1 was knocked by UCA1-siRNA, CDDP chemoresistance weakened, suggesting that UCA1 facilitated proliferation, restrained apoptosis and conferred CDDP resistance of OSCC cells. Luciferase reporter assay showed UCA1, as a ceRNA, downregulated its expression and upregulated steroidogenic factor-1 (SF-1), an essential regulator of tissue-specific gene expression in steroidogenic cells, via sponging miR-184 in OSCC cells [95]. However, the inter-regulation between UCA1 and miR-184 needed intermediate-the store-operated Ca2+ entry (SOCE), but its role between the two were unknown [152].
Interestingly, AC132217.4 was another upregulated lncRNA in OSCC samples. Additionally, krüppel-like factor 8 (KLF8) and insulin-like growth factor 2 (IGF2) have also been overexpressed. KLF8, one of the krüppel-like C2H2 zinc-finger transcription factor family proteins, was considered to exert roles in cancer initiation and progression [153, 154]. IGF2 was an anti-apoptotic endocrine protein, and its upregulation existed in many cancers [155]. Elevated serum IGF2 was proven to be associated with increased risk of developing various cancers including colorectal, prostate and lung [156]. But the regulatory mechanisms between KLF8 and IGF2 were still unknown. AC132217.4 could upregulate IGF2 levels by interacting with 3’UTR of IGF2 mRNA. In addition, cell migration and EMT are promoted [96]. It found that transcription factor STAT3 could positively regulate HNF1A-AS1 levels, and Notch1 and Hes1 (the core factors of Notch signaling pathway) interacting with STAT3 could upregulate HNF1A-AS1 to accelerate OSCC progression [97]. HAS2-AS1 could stabilize HAS2 to promote hypoxia-induced EMT of OSCC cells [98].
HOTAIR regulated E-cadherin through binding oncogene enhancer of zeste homolog 2 and H3K27me3, and it had a negative association with E-cadherin [99]. E-cadherin, expressed in most epithelial cells, was a calcium-dependent transmembrane glycoprotein, and it decreased in patients with OSCC [157, 158]. HOTAIR deletion resulted in downregulating expression of MAP 1LC3B (microtubule-associated protein 1 light chain 3B), beclin1 and autophagy-related gene (ATG) 3, and then autophagy was inhibited. The proliferation and metastasis ability of OSCC cells was also correspondingly weakened [54]. Regarding autophagy, these three molecules each play a complementary function. MAP 1LC3B, an essential protein for autophagosome elongation, were elevated in the tumor tissues of three subsites [159]. Beclin-1 may regulate autophagy process by forming the beclin-1 interactome with some co-factors such as Class III phosphatidylinositol 3-kinase (PI3KCIII)/Vps34, Vps15 [159]. ATG 3 belonged to autophagy-related proteins (ATGs) that regulated the autophagy process in the body [160].
Linc-RoR was first reported to be overexpressed in OSCC tumor specimens [101]. Meanwhile, downregulation of miR-145-5p and overexpression of c-Myc, Klf4, Oct4 and Sox2 indicated the existence of linc-RoR-mediated regulatory network [101]. MiR-145–5p, downregulated in several tumors, was a one of the crucial tumor suppressors, and be proposed as an important regulator of Sox2 [161]. It further suggested that linc-RoR and CCAT1 may share a partially coincident signaling pathway. OSCC tumorigenesis was deteriorated by overexpressed LINC00668 via miR-297/VEGFA axis. However, the mechanism was still to be further clarified [102]. Zhang et al. [104] found that lncRNA ferritin heavy chain 1 pseudogene 3 (FTH1P3) could serve as a molecular sponge of miR-224-5p to modulate fizzled 5 expression, an oncogene in OSCC cells, and facilitate OSCC progression. LINC00152 was found to be at a higher level in OSCC patient tissues. To further explore the role of LINC00152 in OSCC cells, Li et al. [162] transfected SCC9 cells with the sh-LINC00152 plasmid to decrease LINC00152 levels, and LINC00152 knockdown inhibited the proliferation of SCC-9 cells in turn. Moreover, cell proliferation, colony formation, migration, invasion, and the epithelial to mesenchymal transition were inhibited in vitro, as well as tumor growth was disturbed in vivo. A negative correlation between LINC00152 and miR-139 levels indicated LINC00152 acted as a miRNA sponge for miR-139-5p in OSCC [162]. Their targets are stated in the Table 3 [70, 88,89,90,91,92,93,94,95,96, 99,100,101,102,103,104,105].
By using bioinformatic analysis, it had been confirmed that there were 160 differentially expressed lncRNAs between OSCC and normal controls. Moreover, lncRNA FTH1P3, PDIA3F and GTF2IRD2P1 affected the progression and metastasis of OSCC by triggering MMP1, MMP3, MMP9, PLAU and IL8 [163]. There were 21 lncRNAs were significantly related to overall survival (OS) and disease-free survival (DFS) [164]. Among these 21 lncRNAs, a significant positive correlation was observed between the signatures of 13 lncRNAs (TTC39A-AS1, RP11-93B14.9, AC012456.4, RP11-87C12.5, RP11-464F9.21, LINC01549, RP11-897 M7.1, AP003900.6, LINC01343, RP11-181E10.3, CTD-2545H1.2, RP11-796E2.4 and LINC01108) and OS/DFS, while the signatures of the remaining eight lncRNAs (AC007879.2, BOK-AS1, CTB-161 M19.4, CTD-2033A16.3, FAM95B1, RP11-1C8.7, RP11-285G1.14 and RP11-286E11.1) were significantly negatively correlated with OS and DFS [164]. The above signal path can be seen in Fig. 5.
LncRNAs trigger OSCC through multiple regulatory signaling pathways. A variety of methods have been used to detect upregulation or downregulation of multiple lncRNAs in OSCC patients. These lncRNAs are affected by upstream regulatory factors or affect downstream factors to trigger carcinogenic or tumor suppressor signaling pathways
Potential clinical application of lncRNA in OSCC as biomarker
Numerous lncRNAs were aberrantly expressed in various cancers, and some lncRNAs seemed to have been more cancer-specific. Most of them were stable in body fluids and detectable in the plasma and urine of cancer patients. Their expression levels were indicative of the severity of cancers. All of these factors contributed to lncRNAs as noninvasive biomarkers and therapeutic targets for treatment of cancers [165]. LncRNAs were different from protein-coding genes in many respects. First, due to their greater abundance than protein-coding genes, a modulation in a larger number of lncRNA expression may be observed in each subtype of cancer, which provided a larger window for the detection of subtype-specific lncRNA-based biomarker. Second, subtype/tissue–specific lncRNA expressions were crucial for developing novel diagnostic biomarker and personalized therapy [166, 167]. Furthermore, given their participation in diverse cellular signaling pathways and tissue-specific expression, lncRNAs can be utilized to formulate novel strategies for specific cancer subtype diagnosis and targeting. The effects of several representative lncRNAs are shown in the Table 4.
Some lncRNAs were already implicated as biomarkers, though some of them were in clinical trials (Table 5) [36, 103, 130, 137, 168]. For example, compared with the adjacent normal tissues, lncRNA C5orf66-AS1 expression was significantly decreased in OSCC tissues. LncRNA EGFR-AS1 was highly upregulated in the neck squamous cell carcinoma, and was speculated to be an OSCC biomarker [169]. Thus, lncRNAs appeared to be promising novel diagnostic and prognostic markers for a variety of cancers, however, there were still many challenges and validations required for their clinical applications. The utility of circulating and salivary lncRNAs as potential biomarkers gained interest in oral cancers. Plasma levels of HOTAIR and other two lncRNAs (lincRNA-p21 and GAS5) were measured by quantitative polymerase chain reaction, and it found that they were associated with the treatment response of 41 patients with head and neck cancer who underwent radical chemo radio therapy [170]. Maarabouni et al. [171] found higher expression of GAS5 in the patients with progressive disease when compared with those with the good clinical responses. Blood and saliva may provide novel insights into the establishment of new protocols for the detection patients with OSCC.
Multiple lncRNAs were demonstrated to have tumorigenic effects via emerging technologies. LncRNAs, being large in size, may fold into complex secondary/tertiary structures and scaffolds, which may aid in cancer initiation and progression. Li et al. [172] disclosed the ceRNA network and indicated that two lncRNAs (PART1, TTTY14), four miRNAs (hsa-mir-133a, hsa-mir-135b, hsa-mir-196b, hsa-mir193b) and one transcription factor (MEIS1) might be closely related to OSCC tumorigenesis. After DNA damage in Tca8113 cell, HOTAIR mRNA expression increased, and further promoted Tca8113 cell proliferation. When HOTAIR mRNA expression was affected, Tca8113 cell proliferation was blocked in the G2/M or M phase [168]. This indicated that HOTAIR had an oncogenic role and might be an eligible target for OSCC treatment. OSCC patients were found to express higher levels of lncRNA-FOXCUT (a new lncRNA FOXC1 upstream transcript) and mRNA-FOXC1 (fork head box C1 gene) by RT-PCR detection. FOXCUT level was downregulated by siRNA, and FOXC1 level was also downregulated, indicating that FOXCUT was a regulator of FOXC1. Downregulation of FOXCUT and FOXC1 levels inhibited the expression of MMPs (preventing proliferation and migration of OSCC cell (Tca8113 and SCC-9)) and angiogenesis factor VEGF-A (blocking OSCC angiogenesis) [173, 174]. It also revealed that UCA1 had an oncogenic role in OSCC cells in vivo and in vitro [94].
Increased NEAT1 (nuclear paraspeckle assembly transcript 1) levels in OSCC tissues and cells were consistent with advanced TNM stage and poor survival of patients. High levels of NEAT1 antagonized miR-365 (a potential tumor suppressor or oncogene) expressions. Meanwhile, downregulation of NEAT1 levels inhibited cell proliferation and infiltration, suggesting that OSCC could be treated by modulating NEAT1/miR-365 levels [103, 175]. High expression levels of H19 in OSCC tissues were also found to be associated with TNM stage, nodal invasion and a shorter OS. Low expression of H19 can interfere with the proliferation of OSCC cell and inhibit tumor growth [176]. Yu et al. [177] demonstrated that the OS of low and high expression LINC00152 groups were 35 and 28 months, and the RFS were 29 and 26.5 months. There results indicated that LINC00152 may serve as an oncogene in OSCC, and might be a biomarker for early detection, treatment and prognosis prediction of OSCC. An expanded case-control study found that abnormal AC007271.3 levels were significantly associated with clinical stage of OSCC. It suggested that AC007271.3 could be novel circulating biomarkers for the determination of OSCC [178].
In addition to tumorigenic effects, some lncRNAs also played a role in inhibiting tumors. Yang et al. [179] found that GAS5 content in OSCC was lower than that in normal tissues, and suggested that the overexpression of GAS5 inhibited tumor proliferation, migration and invasion ability. Therefore, GAS5 may be clinically used as an anti-oncogene and provided a new target for the treatment of OSCC. Meanwhile, targeted knockout of HOTAIR can be used as a method of treating OSCC [100].
LncRNAs in OSCC prognosis
Surgical resection was considered to be a promising treatment strategy for cancer patients at the early stages. However, recurrence after surgical resection was still a major cause of OSCC-related death. According to report, location, risk factors, clinical stage and treatment, etc. may affect the prognosis of OSCC patients [180]. Biomarkers that predicted the prognosis of patients early were urgently needed to be clinically demonstrated, but they were still not ideal.
Because of the unique role of lncRNAs in OSCC patients, the researchers turned their attention to these molecules. Dong et al. [105] found plasma levels of lncRNA CASC2 decreased in patients with local recurrence but increased in patients without recurrence. And lncRNA CASC2 overexpression promoted cancer cell proliferation. Therefore, CASC2 may participate in the prognosis of OSCC after surgical resection. FLJ22447, referred to LncRNA-CAF, was first found that upregulated CAF was associated with poor prognosis, suggesting it acts as a novel potential OSCC therapeutic target [106]. High expression of HNF1A-AS1 in OSCC samples suggested a poor prognosis, while HNF1A-AS1 deletion inhibited the proliferation, migration and EMT of OSCC cells [97]. Zhou et al. [88] found high MALAT1 levels in 54 OSCC tumor samples and individuals accompanied the poor prognosis. LINC01133 was downregulated in OSCC; higher expression of LINC01133 in OSCC was correlated with less metastasis and better prognosis [181].
The undifferentiated OSCC exhibited a high linc-RoR expression. The phenomenon can be attributed to linc-RoR overexpression interacting with miR-145, causing increased pluripotent transcription factors that regulated the cellular differentiation. High levels of linc-RoR were detected in tissue samples from tumor relapse and drug-resistant patients, suggesting that clinical detection of linc-RoR level predicted the prognosis and therapeutic effects of OSCC [101]. Furthermore, result of the data in the TCGA database concluded that CCAT1 overexpression was associated with poor survival, suggesting that high levels of CCAT1 presented poor therapeutic responses [90]. The expression levels of SOX21-AS1 in OSCC cells were significantly reduced when compared with adjacent normal tissues. In addition, the data showed that the low expression level of SOX21-AS1 was associated with an advanced stage (P = 0.047), large tumor size (P = 0.033), and poor survival in OSCC patients (P = 0.002). These results suggested that low levels of SOX21-AS1 expression may indicate the poor prognosis in OSCC patients [182]. Multivariate Cox proportional hazards regression analyses were used to further determine AC012456.4 low expression as an independent prognostic risk factor (DFS: P = 0.004, HR = 0.600, 95% CI = 0.423–0.851; OS: P = 0.002, HR = 0.672, 95% CI = 0.523–0.863). Moreover, AC012456.4 was pointed out for the first time that it can be used as a novel molecular target for clinical diagnosis, treatment and prognosis for OSCC patients [164].
These lncRNAs work together to form a complex regulatory network, and the credibility of the prognosis is predictive. It is urgent to explore one or a group of lncRNAs as a prognostic indicator in the future.
Future expectations
Considering that lncRNAs have high cell-type specificity, they have been utilized for selectively kill tumor cells without damaging normal cells. H19-DTA (BC-819), a DNA plasmid that carries the gene for diphtheria toxin-A, is used to target H19 overexpressing cancer cells under the regulation of the H19 promoter sequence. The injection of H19-DTA reduces the size of multiple tumor types by inducing the expression of diphtheria toxin [183]. Lavie et al. [184] conducted a phase 1-2A multi-centric trial included 14 eligible ovarian/peritoneal cancer patients. H19-DTA was injected into patients by intra-peritoneal instillations for a maximum of 6–9 weeks. During the study, no dose-limiting toxicities were observed and median survivals of 3.2, 5.3 and 6.5 months were observed for the 60, 120 and 240 mg cohorts, respectively. These results indicated that H19-DTA given locally can provide ancillary therapeutic effects for systemic chemotherapy in ovarian or peritoneal cancer. Gofrit rt. al. [185] included 47 patients with recurrent, multiple nonmuscle invasive bladder tumors in a phase 2b trial. Patients expressing H19 received a 6-week induction course of intravesical H19-DTA. 33% of patients showed complete tumor ablation and 64% were no new tumors at 3 months. At the same time, the median time to recurrence was also significantly prolonged in responding patients. It was concluded that H19-DTA served as a potential medication for bladder cancer. These results are gratifying. Because it allows us to speculate that other lncRNAs have similar therapeutic effects, although they have not been discovered at this stage.
LncRNA research in OSCC was also still incipient. The upregulated and downregulated lncRNA profiles in oral cancers should be established [186], but the differentially expressed lncRNAs needed to be functionally evaluated in the context of the cells investigated. Meanwhile, the road to exploration was not going well. For example, many lncRNAs are located in the nucleus and are difficult to knock out [187].
Recently, the successful application of CRISPR-Cas9 (clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9) technology to model plants has given us new inspiration. CRISPR-Cas9 is considered to be a bacterial defense mechanism against phage infection and plasmid transfer and simple, versatile and promising genome editing technique [188]. Use of CRISPR-Cas9 for the treatment of schizophrenia has once again enhanced our confidence in treating cancer [189]. The lncRNAs editing protocol based on CRISPR-Cas9 technology has been widely used in a variety of diseases, including cancer, albeit there are currently limitations in this system [190]. There is not enough research to prove that Cas9 technology can be applied to OSCC, but some theoretical connections let us see hope [191]. This technology is expected to use to treat OSCC and clarify the relevant mechanisms.
Exosomes, nanosized (30-100 nm) membrane microvesicles, can act as messengers in the interstitial to establish communication between cancer cells and basal cells [192]. There is evidence that exosomes regulate tumor growth and metastasis through inclusions, containing lncRNAs (Table 6) [193,194,195,196,197,198,199,200,201,202,203], and serve as noninvasive biomarkers for early detection, diagnosis, and treatment of cancer patients [204]. Zhang et al. [193] found that the MALAT-1 content in exosomes of lung cancer patients was overexpressed. Functionally, serum exosome-derived MALAT-1 promoted tumor growth and metastasis. It indicated that MALAT-1 in exosomes can be used as a noninvasive biomarker for diagnosis and prognosis of non-small cell lung cancer [193]. Therefore, finding out the specific lncRNAs in the exosomes of patients with OSCC is the direction of future exploration.
Conclusion
It is imperative to crack the carcinogenic or tumor suppressor mechanism of lncRNAs, which is of great significance for the diagnosis and treatment of cancer by utilizing lncRNAs. A curated collection and summary of deregulated lncRNAs in cancer is essential to thoroughly understand the mechanisms and functions of lncRNAs. The low accessible amount of lncRNAs poses an obstacle to detection. At the same time, the mechanism of interaction between lncRNAs is poorly understood. Problems such as these are believed to be solved using animal models and new generation of technologies. LncRNA studies in oral cancer are expected to undergo a vast expansion in decades. Furthermore, we suggest strategies to accelerate the pace from the bench to the bedside.
Abbreviations
- 3-UTRs:
-
3-untranslated regions
- ATG:
-
Autophagy-related gene
- CCAT1:
-
Colon Cancer Associated Transcript 1
- CDKN2A:
-
Cyclin-dependent kinase inhibitor 2a
- ceRNA:
-
competing endogenous RNA
- circRNAs:
-
circular RNAs
- CRISPR-Cas9:
-
Clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9
- DFS:
-
Disease-free survival
- DLEU1:
-
Deleted in lymphocytic leukemia 1
- DPD:
-
Dihydropyrimidine dehydrogenase
- EGFR:
-
Epidermal growth factor receptor
- EMT:
-
Epithelial-mesenchymal transition
- FOXC1:
-
Fork head box C1 gene
- FTH1P3:
-
Ferritin heavy chain 1 pseudogene 3
- Gtl2:
-
Gene trap locus2
- HIOECs:
-
Human immortalized oral epithelial cells
- HOTAIR:
-
HOX transcription antisense RNA
- HPV:
-
Human papillomavirus
- lncRNAs:
-
long non-coding RNAs
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- MAP 1 LC3B:
-
Microtubule-associated protein 1 light chain 3B
- MEG3:
-
Maternally expressed gene 3
- miRNAs:
-
microRNAs
- ncRNAs:
-
non-coding RNAs
- NOTCH1:
-
Notch homolog 1 genes are translocation-associated
- OS:
-
Overall survival
- OSCC:
-
Oral squamous cell carcinoma
- P. gingivalis :
-
Porphyromonas gingivalis
- STAT3:
-
Signal transducer and activator of transcription 3
- UCA1:
-
Urothelial cancer associated 1
References
Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma--an update. CA Cancer J Clin. 2015;65(5):401–21.
Pannone G, Santoro A, Papagerakis S, Lo Muzio L, De Rosa G, Bufo P. The role of human papillomavirus in the pathogenesis of head & neck squamous cell carcinoma: an overview. Infect Agent Cancer. 2011;6(1):4.
Sankunny M, Parikh RA, Lewis DW, Gooding WE, Saunders WS, Gollin SM. Targeted inhibition of ATR or CHEK1 reverses radioresistance in oral squamous cell carcinoma cells with distal chromosome arm 11q loss. Genes Chromosomes Cancer. 2014;53(2):129–43.
Mehrotra R, Yadav S. Oral squamous cell carcinoma: etiology, pathogenesis and prognostic value of genomic alterations. Indian J Cancer. 2006;43(2):60–6.
Patel SC, Carpenter WR, Tyree S, Couch ME, Weissler M, Hackman T, Hayes DN, Shores C, Chera BS. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol. 2011;29(11):1488–94.
Feller LL, Khammissa RR, Kramer BB, Lemmer JJ. Oral squamous cell carcinoma in relation to field precancerisation: pathobiology. Cancer Cell Int. 2013;13(1):31.
Curry JM, Sprandio J, Cognetti D, Luginbuhl A, Bar-ad V, Pribitkin E, Tuluc M. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol. 2014;41(2):217–34.
Yuan Z, Yu Y, Zhang B, Miao L, Wang L, Zhao K, Ji Y, Wang R, Ma H, Chen N, et al. Genetic variants in lncRNA H19 are associated with the risk of oral squamous cell carcinoma in a Chinese population. Oncotarget. 2018;9(35):23915–22.
Gandini S, Botteri E, Iodice S, Boniol M, Lowenfels AB, Maisonneuve P, Boyle P. Tobacco smoking and cancer: a meta-analysis. Int J Cancer. 2008;122(1):155–64.
Lee J, Taneja V, Vassallo R. Cigarette smoking and inflammation: cellular and molecular mechanisms. J Dent Res. 2012;91(2):142–9.
Yamashita T, Kato K, Long NK, Makita H, Yonemoto K, Iida K, Tamaoki N, Hatakeyama D, Shibata T. Effects of smoking and alcohol consumption on 5-fluorouracil-related metabolic enzymes in oral squamous cell carcinoma. Molecular and clinical oncology. 2014;2(3):429–34.
Guo LK, Zhang CX, Guo XF. Association of genetic polymorphisms of aldehyde dehydrogenase-2 and cytochrome P450 2E1-RsaI and alcohol consumption with oral squamous cell carcinoma. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2012;34(4):390–5.
Ramqvist T, Grun N, Dalianis T. Human papillomavirus and tonsillar and base of tongue cancer. Viruses. 2015;7(3):1332–43.
Bloebaum M, Poort L, Böckmann R, Kessler P. Survival after curative surgical treatment for primary oral squamous cell carcinoma. J Craniomaxillofac Surg. 2014;42(8):1572–6.
Kummerow KL, Du L, Penson DF, Shyr Y, Hooks MA. Nationwide trends in mastectomy for early-stage breast cancer. JAMA Surg. 2015;150(1):9–16.
Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–89.
Araldi E, Suárez Y. MicroRNAs as regulators of endothelial cell functions in cardiometabolic diseases. Biochim Biophys Acta. 2016;1861(12 Pt B):2094–103.
Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–46.
Delás MJ, Sabin LR, Dolzhenko E, Knott SR, Munera Maravilla E, Jackson BT, Wild SA, Kovacevic T, Stork EM, Zhou M, et al. lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. eLife. 2017;6:e25607.
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69.
Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18.
Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253.
Schmitt AM, Chang HY. Long noncoding RNAs in Cancer pathways. Cancer Cell. 2016;29(4):452–63.
Wu J, Xie H. Expression of long noncoding RNA-HOX transcript antisense intergenic RNA in oral squamous cell carcinoma and effect on cell growth. Tumour Biol. 2015;36(11):8573–8.
Hema KN, Smitha T, Sheethal HS, Mirnalini SA. Epigenetics in oral squamous cell carcinoma. J Oral Maxillofac Pathol. 2017;21(2):252–9.
Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, Henson B, Hu S, Arellano M, Sinha U, Le A, et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomark Prev. 2012;21(4):664–72.
Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7(3):761–6.
Huang SH, O'Sullivan B. Oral cancer: current role of radiotherapy and chemotherapy. Med Oral Patol Oral Cir Bucal. 2013;18(2):e233–40.
D'Cruz AK, Vaish R, Dhar H. Oral cancers: current status. Oral Oncol. 2018;87:64–9.
Gharat SA, Momin M, Bhavsar C. Oral squamous cell carcinoma: current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit Rev Ther Drug Carrier Syst. 2016;33(4):363–400.
Yu T, Li C, Wang Z, Liu K, Xu C, Yang Q, Tang Y, Wu Y. Non-coding RNAs deregulation in oral squamous cell carcinoma: advances and challenges. Clin Transl Oncol. 2016;18(5):427–36.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407.
Yang Y, Wen L, Zhu H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015;5(1):59.
Meryet-Figuière M, Lambert B, Gauduchon P, Vigneron N, Brotin E, Poulain L, Denoyelle C. An overview of long non-coding RNAs in ovarian cancers. Oncotarget. 2016;7(28):44719–34.
Sun Q, Hao Q, Prasanth KV. Nuclear Long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–57.
Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37(2):1437–44.
Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3 UTRs via Alu elements. Nature. 2011;470(7333):284–8.
Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136(4):604–9.
Novikova IV, Hennelly SP, Sanbonmatsu KY. Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? Bioarchitecture. 2012;2(6):189–99.
Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151(3):939–47.
Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44(2):863–77.
Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62.
Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14(1):35–42.
Fang S, Lee HJ, Wark AW, Corn RM. Attomole microarray detection of microRNAs by nanoparticle-amplified SPR imaging measurements of surface polyadenylation reactions. J Am Chem Soc. 2006;128(43):14044–6.
Nasheri N, Cheng J, Singaravelu R, Wu P, McDermott MT, Pezacki JP. An enzyme-linked assay for the rapid quantification of microRNAs based on the viral suppressor of RNA silencing protein p19. Anal Biochem. 2011;412(2):165–72.
Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3(1):27–9.
Hanna JA, Wimberly H, Kumar S, Slack F, Agarwal S, Rimm DL. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. Biotechniques. 2012;52(4):235–45.
Tang X, Gal J, Zhuang X, Wang W, Zhu H, Tang G. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13(10):1803–22.
Zhao B, Jin L, Wei J, Ma Z, Jiang W, Ma L, Jin Y. A simple and fast method for profiling microRNA expression from low-input total RNA by microarray. IUBMB Life. 2012;64(7):612–6.
Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SK. Bioluminescence-based detection of microRNA, miR21 in breast cancer cells. Anal Chem. 2008;80(7):2319–25.
Cissell KA, Deo SK. Trends in microRNA detection. Anal Bioanal Chem. 2009;394(4):1109–16.
Peng Y, Gao Z. Amplified detection of microRNA based on ruthenium oxide nanoparticle-initiated deposition of an insulating film. Anal Chem. 2011;83(3):820–7.
Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–16.
Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, Tang J, Chen X, Dai J, Wei Q, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33(4):828–34.
Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndic M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. 2010;5(11):807–14.
Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat Nanotechnol. 2011;6(10):668–74.
Jensen SG, Lamy P, Rasmussen MH, Ostenfeld MS, Dyrskjot L, Orntoft TF, Andersen CL. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 2011;12:435.
Bianchi N, Zuccato C, Finotti A, Lampronti I, Borgatti M, Gambari R. Involvement of miRNA in erythroid differentiation. Epigenomics. 2012;4(1):51–65.
Robertson KL, Vora GJ. Locked nucleic acid and flow cytometry-fluorescence in situ hybridization for the detection of bacterial small noncoding RNAs. Appl Environ Microbiol. 2012;78(1):14–20.
Sioss JA, Bhiladvala RB, Pan W, Li M, Patrick S, Xin P, Dean SL, Keating CD, Mayer TS, Clawson GA. Nanoresonator chip-based RNA sensor strategy for detection of circulating tumor cells: response using PCA3 as a prostate cancer marker. Nanomedicine. 2012;8(6):1017–25.
Jiang L, Duan D, Shen Y, Li J. Direct microRNA detection with universal tagged probe and time-resolved fluorescence technology. Biosens Bioelectron. 2012;34(1):291–5.
Alhasan AH, Kim DY, Daniel WL, Watson E, Meeks JJ, Thaxton CS, Mirkin CA. Scanometric microRNA array profiling of prostate cancer markers using spherical nucleic acid-gold nanoparticle conjugates. Anal Chem. 2012;84(9):4153–60.
Dong H, Zhang J, Ju H, Lu H, Wang S, Jin S, Hao K, Du H, Zhang X. Highly sensitive multiple microRNA detection based on fluorescence quenching of graphene oxide and isothermal strand-displacement polymerase reaction. Anal Chem. 2012;84(10):4587–93.
Li R, Zhu H, Luo Y. Understanding the functions of Long non-coding RNAs through their higher-order structures. Int J Mol Sci. 2016;17(5):702.
Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486.
Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc. 2015;10:1643.
Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):924–33.
Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, Gao Y, Guo M, Yue M, Wang L, et al. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016;44(D1):D980–5.
Zou Y, Li C, Shu F, Tian Z, Xu W, Xu H, Tian H, Shi R, Mao X. lncRNA expression signatures in periodontitis revealed by microarray: the potential role of lncRNAs in periodontitis pathogenesis. J Cell Biochem. 2015;116(4):640–7.
Nishiyama K, Maruyama R, Niinuma T, Kai M, Kitajima H, Toyota M, Hatanaka Y, Igarashi T, Kobayashi J-I, Ogi K, et al. Screening for long noncoding RNAs associated with oral squamous cell carcinoma reveals the potentially oncogenic actions of DLEU1. Cell Death Dis. 2018;9(8):826.
Yang Y, Wang Y, Lai J, Shen S, Wang F, Kong J, Zhang W, Yang H. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/β-catenin signaling pathway. Cancer Sci. 2016;107(11):1581–9.
Jun T, Zheng FS, Ren KM, Zhang HY, Zhao JG, Zhao JZ. Long non-coding RNA UCA1 regulates the proliferation, migration and invasion of human lung cancer cells by modulating the expression of microRNA-143. Eur Rev Med Pharmacol Sci. 2018;22(23):8343–52.
Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y, Zhou M, Fan H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018;9(12):1158.
Luo J, Chen J, Li H, Yang Y, Yun H, Yang S, Mao X. LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett. 2017;14(5):5556–62.
Chen L-L, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22(3):357–64.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14.
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24(5):513–31.
Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, Cui J, Wang G, Qian G, Higgins MJ, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol. 2014;204(1):61–75.
Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–20.
Xu T, CM L, Cheng SQ, Min J, Li L, Meng XM, Huang C, Zhang L, Deng ZY, Li J. Pathological bases and clinical impact of long noncoding RNAs in prostate cancer: a new budding star. Mol Cancer. 2018;17(1):103.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344.
Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75–93.
Wang T, Yuan J, Feng N, Li Y, Lin Z, Jiang Z, Gui Y. Hsa-miR-1 downregulates long non-coding RNA urothelial cancer associated 1 in bladder cancer. Tumour Biol. 2014;35(10):10075–84.
Muller S, Raulefs S, Bruns P, Afonso-Grunz F, Plotner A, Thermann R, Jager C, Schlitter AM, Kong B, Regel I, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94.
Koduru SV, Tiwari AK, Leberfinger A, Hazard SW, Kawasawa YI, Mahajan M, Ravnic DJ. A comprehensive NGS data analysis of differentially regulated miRNAs, piRNAs, lncRNAs and sn/snoRNAs in triple negative breast Cancer. J Cancer. 2017;8(4):578–96.
Hon KW, Abu N, Ab Mutalib NS. Jamal R miRNAs and lncRNAs as Predictive Biomarkers of Response to FOLFOX Therapy in Colorectal Cancer. Front Pharmacol. 2018;9:846.
Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65(6):1140–51.
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren Y, Wu Y, Mei M, Zhang L, Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in Oral squamous cell carcinoma. Sci Rep. 2015;5:15972.
Chang SM, Hu WW. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J Cell Physiol. 2018;233(4):3384–96.
Arunkumar G, Murugan AK. Prasanna Srinivasa Rao H, Subbiah S, Rajaraman R, Munirajan AK. Long non-coding RNA CCAT1 is overexpressed in oral squamous cell carcinomas and predicts poor prognosis. Biomedical Reports. 2017;6(4):455–62.
Geng F, Liu J, Guo Y, Li C, Wang H, Wang H, Zhao H, Pan Y. Persistent exposure to Porphyromonas gingivalis promotes proliferative and invasion capabilities, and tumorigenic properties of human immortalized Oral epithelial cells. Front Cell Infect Microbiol. 2017;7:57.
Shiah SG, Hsiao JR, Chang WM, Chen YW, Jin YT, Wong TY, Huang JS, Tsai ST, Hsu YM, Chou ST, et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014;74(24):7560–72.
Liu Z, Wu C, Xie N, Wang P. Long non-coding RNA MEG3 inhibits the proliferation and metastasis of oral squamous cell carcinoma by regulating the WNT/beta-catenin signaling pathway. Oncol Lett. 2017;14(4):4053–8.
Yang YT, Wang YF, Lai JY, Shen SY, Wang F, Kong J, Zhang W, Yang HY. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/beta-catenin signaling pathway. Cancer Sci. 2016;107(11):1581–9.
Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. 2017;6(12):2897–908.
Li X, Ma C, Zhang L, Li N, Zhang X, He J, He R, Shao M, Wang J, Kang L, et al. LncRNAAC132217.4, a KLF8-regulated long non-coding RNA, facilitates oral squamous cell carcinoma metastasis by upregulating IGF2 expression. Cancer Lett. 2017;407:45–56.
Liu Z, Li H, Fan S, Lin H, Lian W. STAT3-induced upregulation of long noncoding RNA HNF1A-AS1 promotes the progression of oral squamous cell carcinoma via activating notch signaling pathway. Cancer Biol Ther. 2018;1:10.
Zhu G, Wang S, Chen J, Wang Z, Liang X, Wang X, Jiang J, Lang J, Li L. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol Carcinog. 2017;56(10):2210–22.
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, Wei F, Yu W, Liu T, Wang X, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol. 2015;46(6):2586–94.
Wang X, Liu W, Wang P, Li S. RNA interference of long noncoding RNA HOTAIR suppresses autophagy and promotes apoptosis and sensitivity to cisplatin in oral squamous cell carcinoma. J Oral Pathol Med. 2018;47(10):930–7.
Arunkumar G, Deva Magendhra Rao AK, Manikandan M, Arun K, Vinothkumar V, Revathidevi S, Rajkumar KS, Rajaraman R, Munirajan AK. Expression profiling of long non-coding RNA identifies linc-RoR as a prognostic biomarker in oral cancer. Tumour Biol. 2017;39(4):1010428317698366.
Zhang CZ. Long intergenic non-coding RNA 668 regulates VEGFA signaling through inhibition of miR-297 in oral squamous cell carcinoma. Biochem Biophys Res Commun. 2017;489(4):404–12.
Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2):e12329.
Zhang CZ. Long non-coding RNA FTH1P3 facilitates oral squamous cell carcinoma progression by acting as a molecular sponge of miR-224-5p to modulate fizzled 5 expression. Gene. 2017;607:47–55.
Dong Y, Wu W. Downregulation of lncRNA CASC2 promotes the postoperative local recurrence of early oral squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2019;276(2):605–10.
Ding L, Ren J, Zhang D, Li Y, Huang X, Hu Q, Wang H, Song Y, Ni Y, Hou Y. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis. 2018;39(3):397–406.
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7.
Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–9.
Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39.
Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–32.
Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A. 2012;109(47):19202–7.
Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–38.
Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3(2)
Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–14.
Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–88.
Xu S, Sui S, Zhang J, Bai N, Shi Q, Zhang G, Gao S, You Z, Zhan C, Liu F, et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8(5):4881–91.
Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39(1):169–75.
Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368.
Ertosun MG, Hapil FZ, Osman NO. E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine Growth Factor Rev. 2016;31:17–25.
Tano K, Mizuno R, Okada T, Rakwal R, Shibato J, Masuo Y, Ijiri K, Akimitsu N. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–80.
Musa J, Aynaud MM, Mirabeau O, Delattre O, Grunewald TG. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017;8(6):e2895.
Onoue T, Uchida D, Begum NM, Tomizuka Y, Yoshida H, Sato M. Epithelial-mesenchymal transition induced by the stromal cell-derived factor-1/CXCR4 system in oral squamous cell carcinoma cells. Int J Oncol. 2006;29(5):1133–8.
Henson BJ, Bhattacharjee S, O'Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48(7):569–82.
Sarode GS, Sarode SC, Patil A, Anand R, Patil SG, Rao RS, Augustine D. Inflammation and Oral Cancer: An update review on targeted therapies. J Contemp Dent Pract. 2015;16(7):595–602.
Liu LH, Li H, Li JP, Zhong H, Zhang HC, Chen J, Xiao T. miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem Biophys Res Commun. 2011;416(1–2):31–8.
He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35(12):12181–8.
Perez-Sayans M, Suarez-Penaranda JM, Pilar GD, Barros-Angueira F, Gandara-Rey JM, Garcia-Garcia A. What real influence does the proto-oncogene c-myc have in OSCC behavior? Oral Oncol. 2011;47(8):688–92.
Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells. 2016;39(4):330–6.
Lu L, Xu H, Luo F, Liu X, Lu X, Yang Q, Xue J, Chen C, Shi L, Liu Q. Epigenetic silencing of miR-218 by the lncRNA CCAT1, acting via BMI1, promotes an altered cell cycle transition in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2016;304:30–41.
Xin Y, Li Z, Shen J, Chan MT, Wu WK. CCAT1: a pivotal oncogenic long non-coding RNA in human cancers. Cell Prolif. 2016;49(3):255–60.
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583.
Baba O, Hasegawa S, Nagai H, Uchida F, Yamatoji M, Kanno NI, Yamagata K, Sakai S, Yanagawa T, Bukawa H. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J Oral Pathol Med. 2016;45(4):248–55.
Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88(11):5119–26.
Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10(11):1711–8.
Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, Stratakis CA, Hedley-Whyte ET, Klibanski A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J Clin Endocrinol Metab. 2008;93(10):4119–25.
da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24(6):306–16.
Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):R45–53.
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461.
Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743.
Jia LF, Wei SB, Gan YH, Guo Y, Gong K, Mitchelson K, Cheng J, Yu GY. Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma. Int J Cancer. 2014;135(10):2282–93.
Bova RJ, Quinn DI, Nankervis JS, Cole IE, Sheridan BF, Jensen MJ, Morgan GJ, Hughes CJ, Sutherland RL. Cyclin D1 and p16INK4A expression predict reduced survival in carcinoma of the anterior tongue. Clin Cancer Res. 1999;5(10):2810–9.
Song JL, Nigam P, Tektas SS, Selva E. microRNA regulation of Wnt signaling pathways in development and disease. Cell Signal. 2015;27(7):1380–91.
Jiang Q, He M, Guan S, Ma M, Wu H, Yu Z, Jiang L, Wang Y, Zong X, Jin F, et al. MicroRNA-100 suppresses the migration and invasion of breast cancer cells by targeting FZD-8 and inhibiting Wnt/beta-catenin signaling pathway. Tumour Biol. 2016;37(4):5001–11.
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014;5:e1008.
Xue M, Chen W, Li X. Urothelial cancer associated 1: a long noncoding RNA with a crucial role in cancer. J Cancer Res Clin Oncol. 2016;142(7):1407–19.
Yang C, Li X, Wang Y, Zhao L, Chen W. Long non-coding RNA UCA1 regulated cell cycle distribution via CREB through PI3-K dependent pathway in bladder carcinoma cells. Gene. 2012;496(1):8–16.
Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol BioSyst. 2015;11(7):1946–54.
Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR Axis. Curr Top Microbiol Immunol. 2017;407:153–89.
Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–27.
Li J, Gao J, Kan A, Hao T, Huang L. SNHG and UCA1 as prognostic molecular biomarkers in hepatocellular carcinoma: recent research and meta-analysis. Minerva Med. 2017;108(6):568–74.
Harada K, Ferdous T, Harada T, Takenawa T, Ueyama Y. Gimeracil enhances the antitumor effect of cisplatin in oral squamous cell carcinoma cells in vitro and in vivo. Oncol Lett. 2017;14(3):3349–56.
Richardson A, Owens DJ, Ross K. MicroRNA-184 and its lncRNA sponge UCA1 are induced in wounded keratinocytes in a store-operated calcium entry-dependent manner. Br J Dermatol. 2018. https://doi.org/10.1111/bjd.17576.
Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32(11–12):1103–21.
Mehta TS, Monzur F, Zhao J. Determination of nuclear localization signal sequences for Kruppel-like factor 8. Methods Mol Biol. 2010;647:171–86.
Brouwer-Visser J, Huang GS. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 2015;26(3):371–7.
Livingstone C. IGF2 and cancer. Endocr Relat Cancer. 2013;20(6):R321–39.
Liu LK, Jiang XY, Zhou XX, Wang DM, Song XL, Jiang HB. Upregulation of vimentin and aberrant expression of E-cadherin/beta-catenin complex in oral squamous cell carcinomas: correlation with the clinicopathological features and patient outcome. Mod Pathol. 2010;23(2):213–24.
Pannone G, Santoro A, Feola A, Bufo P, Papagerakis P, Lo Muzio L, Staibano S, Ionna F, Longo F, Franco R, et al. The role of E-cadherin down-regulation in oral cancer: CDH1 gene expression and epigenetic blockage. Curr Cancer Drug Targets. 2014;14(2):115–27.
Liu PF, Chang HW, Cheng JS, Lee HP, Yen CY, Tsai WL, Cheng JT, Li YJ, Huang WC, Lee CH, et al. Map 1lc3b and Sqstm1 Modulated Autophagy for Tumorigenesis and Prognosis in Certain Subsites of Oral Squamous Cell Carcinoma. J Clin Med. 2018;7(12).
Murrow L, Malhotra R, Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol. 2015;17(3):300–10.
Ozen M, Karatas OF, Gulluoglu S, Bayrak OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ, et al. Overexpression of miR-145-5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. Cancer Investig. 2015;33(6):251–8.
Li M, Ning J, Li Z, Wang J, Zhao C, Wang L. LINC00152 promotes the growth and invasion of oral squamous cell carcinoma by regulating miR-139-5p. Onco Targets Ther. 2018;11:6295–304.
Zhang S, Tian L, Ma P, Sun Q, Zhang K. GuanchaoWang, Liu H, Xu B. potential role of differentially expressed lncRNAs in the pathogenesis of oral squamous cell carcinoma. Arch Oral Biol. 2015;60(10):1581–7.
Hu X, Qiu Z, Zeng J, Xiao T, Ke Z, Lyu H. A novel long non-coding RNA, AC012456.4, as a valuable and independent prognostic biomarker of survival in oral squamous cell carcinoma. PeerJ. 2018;6:e5307.
Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927–33.
Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5(20):9864–76.
Melo CP, Campos CB, Rodrigues Jde O, Aguirre-Neto JC, Atalla A, Pianovski MA, Carbone EK, Lares LB, Moraes-Souza H, Octacilio-Silva S, et al. Long non-coding RNAs: biomarkers for acute leukaemia subtypes. Br J Haematol. 2016;173(2):318–20.
Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–64.
Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL, Zhang X, Sundaram GM, Tan GS, Chang MM, et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23(10):1167–75.
Fayda M, Isin M, Tambas M, Guveli M, Meral R, Altun M, Sahin D, Ozkan G, Sanli Y, Isin H, et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumour Biol. 2016;37(3):3969–78.
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208.
Li S, Chen X, Liu X, Yu Y, Pan H, Haak R, Schmidt J, Ziebolz D, Schmalz G. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9.
Kong XP, Yao J, Luo W, Feng FK, Ma JT, Ren YP, Wang DL, Bu RF. The expression and functional role of a FOXC1 related mRNA-lncRNA pair in oral squamous cell carcinoma. Mol Cell Biochem. 2014;394(1–2):177–86.
Yu T, Wu Y, Helman JI, Wen Y, Wang C, Li L. CXCR4 promotes oral squamous cell carcinoma migration and invasion through inducing expression of MMP-9 and MMP-13 via the ERK signaling pathway. Mol Cancer Res. 2011;9(2):161–72.
Huang G, He X, Wei XL. lncRNA NEAT1 promotes cell proliferation and invasion by regulating miR365/RGS20 in oral squamous cell carcinoma. Oncol Rep. 2018;39(4):1948–56.
Hong Y, He H, Sui W, Zhang J, Zhang S, Yang D. Long non-coding RNA H1 promotes cell proliferation and invasion by acting as a ceRNA of miR138 and releasing EZH2 in oral squamous cell carcinoma. Int J Oncol. 2018;52(3):901–12.
Yu J, Liu Y, Guo C, Zhang S, Gong Z, Tang Y, Yang L, He Y, Lian Y, Li X, et al. Upregulated long non-coding RNA LINC00152 expression is associated with progression and poor prognosis of tongue squamous cell carcinoma. J Cancer. 2017;8(4):523–30.
Shao T, Huang J, Zheng Z, Wu Q, Liu T, Lv X. SCCA, TSGF, and the Long non-coding RNA AC007271.3 are effective biomarkers for diagnosing Oral squamous cell carcinoma. Cell Physiol Biochem. 2018;47(1):26–38.
Yang M, Xiong X, Chen L, Yang L, Li X. Identification and validation long non-coding RNAs of oral squamous cell carcinoma by bioinformatics method. Oncotarget. 2017;8(64):107469–76.
de Morais EF, Mafra RP, Gonzaga AKG, de Souza DLB, Pinto LP, da Silveira EJD. Prognostic factors of Oral squamous cell carcinoma in young patients: a systematic review. J Oral Maxillofac Surg. 2017;75(7):1555–66.
Kong J, Sun W, Zhu W, Liu C, Zhang H, Wang H. Long noncoding RNA LINC01133 inhibits oral squamous cell carcinoma metastasis through a feedback regulation loop with GDF15. J Surg Oncol. 2018;118(8):1326–34.
Yang CM, Wang TH, Chen HC, Li SC, Lee MC, Liou HH, Liu PF, Tseng YK, Shiue YL, Ger LP, et al. Aberrant DNA hypermethylation-silenced SOX21-AS1 gene expression and its clinical importance in oral cancer. Clin Epigenetics. 2016;8:129.
Mizrahi A, Czerniak A, Levy T, Amiur S, Gallula J, Matouk I, Abu-lail R, Sorin V, Birman T, de Groot N, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69.
Lavie O, Edelman D, Levy T, Fishman A, Hubert A, Segev Y, Raveh E, Gilon M, Hochberg A. A phase 1/2a, dose-escalation, safety, pharmacokinetic, and preliminary efficacy study of intraperitoneal administration of BC-819 (H19-DTA) in subjects with recurrent ovarian/peritoneal cancer. Arch Gynecol Obstet. 2017;295(3):751–61.
Gofrit ON, Benjamin S, Halachmi S, Leibovitch I, Dotan Z, Lamm DL, Ehrlich N, Yutkin V, Ben-Am M, Hochberg A. DNA based therapy with diphtheria toxin-a BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J Urol. 2014;191(6):1697–702.
Zhang X, Lopez-Berestein G, Sood AK, Calin GA. Profiling Long noncoding RNA expression using custom-designed microarray. Methods Mol Biol. 2016;1402:33–41.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21.
Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–29.
Zhuo C, Hou W, Hu L, Lin C, Chen C, Lin X. Genomic editing of non-coding RNA genes with CRISPR/Cas9 ushers in a potential novel approach to study and treat schizophrenia. Front Mol Neurosci. 2017;10:28.
Yang J, Meng X, Pan J, Jiang N, Zhou C, Wu Z, Gong Z. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol. 2018;15(1):35–43.
Pulecio J, Verma N, Mejia-Ramirez E, Huangfu D, Raya A. CRISPR/Cas9-based engineering of the epigenome. Cell Stem Cell. 2017;21(4):431–47.
Ruivo CF, Adem B, Silva M, Melo SA. The biology of Cancer exosomes: insights and new perspectives. Cancer Res. 2017;77(23):6480–8.
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490(2):406–14.
Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16(1):143.
Jahangiri B, Khalaj-Kondori M, Asadollahi E, Sadeghizadeh M. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer SW480 cells by provoking long noncoding RNA UCA1. Journal of cell communication and signaling. 2019;13(1):53–64.
Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143(6):991–1004.
Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7(51):85551–63.
Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A, Fucile C, Richheimer S, Beckham CJ. Expression of the Long non-coding RNA HOTAIR correlates with disease progression in bladder Cancer and is contained in bladder Cancer patient urinary exosomes. PLoS One. 2016;11(1):e0147236.
Gao T, Liu X, He B, Nie Z, Zhu C, Zhang P, Wang S. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018;18:11.
Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155.
Lang HL, Hu GW, Zhang B, Kuang W, Chen Y, Wu L, Xu GH. Glioma cells enhance angiogenesis and inhibit endothelial cell apoptosis through the release of exosomes that contain long non-coding RNA CCAT2. Oncol Rep. 2017;38(2):785–98.
Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS open bio. 2014;4:458–67.
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, et al. Exosome-transmitted lncARSR promotes Sunitinib resistance in renal Cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–68.
Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–15.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Nos. 81700522, 81470003), the fund of Key Project of Natural Science Research Project of Anhui Provincial Department of Education (KJ2018A0203), the Grants for Scientific Research of BSKY (XJ201706), the Project Supported by Anhui Provincial Natural Science Foundation (1508085MH187).
Funding
The National Natural Science Foundation of China (Nos. 81700522, 81470003), from the National Natural Science Foundation of China-for the design of the study.
The fund of Key Project of Natural Science Research Project of Anhui Provincial Department of Education (KJ2018A0203), from the Education Department of Anhui, China-for the collection of data.
The Grants for Scientific Research of BSKY (XJ201706), from Anhui Medical University, China-for the analysis of data.
The Project Supported by Anhui Provincial Natural Science Foundation (1508085MH187), China-for the collection of data.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
LZ, XM and XWZ collected the related literature and drafted the manuscript. DCY and RC participated in the design of the review and drafted the manuscript. LZ and XM revised and edited the manuscript. XM, YJ and TX supervised the review process. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, L., Meng, X., Zhu, Xw. et al. Long non-coding RNAs in Oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol Cancer 18, 102 (2019). https://doi.org/10.1186/s12943-019-1021-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12943-019-1021-3