Abstract
Background
In early 2020, COVID-19 pandemic has mobilized researchers in finding new remedies including repurposing of medicinal plant products focusing on direct-acting antiviral and host-directed therapies. In this study, we performed an in vitro investigation on the standardized Marantodes pumilum extract (SKF7®) focusing on anti-SARS-CoV-2 and anti-inflammatory activities.
Methods
Anti-SARS-CoV-2 potential of the SKF7® was evaluated in SARS-CoV-2-infected Vero E6 cells and SARS-CoV-2-infected A549 cells by cytopathic effect-based assay and RT-qPCR, respectively. Target based assays were performed on the SKF7® against the S1-ACE2 interaction and 3CL protease activities. Anti-inflammatory activity of the SKF7® was evaluated by nitric oxide inhibitory and TLR2/TLR4 receptor blocker assays.
Results
The SKF7® inhibited wild-type Wuhan (EC50 of 21.99 µg/mL) and omicron (EC50 of 16.29 µg/mL) SARS-CoV-2 infections in Vero-E6 cells. The SKF7® also inhibited the wild-type SARS-CoV-2 infection in A549 cells (EC50 value of 6.31 µg/mL). The SKF7® prominently inhibited 3CL protease activity. The SKF7® inhibited the LPS induced-TLR4 response with the EC50 of 16.19 µg/mL.
Conclusions
In conclusion, our in vitro study highlighted anti-SARS-CoV-2 and anti-inflammatory potentials of the SKF7®. Future pre-clinical in vivo studies focusing on antiviral and immunomodulatory potentials of the SKF7® in affecting the COVID-19 pathogenesis are warranted.
Similar content being viewed by others
Background
Global situation of COVID-19 has reached more than 775 million cases including 7 million deaths as recorded in May 2024 [1]. COVID-19 is an infectious disease caused by the SARS-CoV-2 viral infection that can be spreads from infected persons through a respiratory droplet to smaller aerosols. COVID-19 patients can experience presymptomatic and asymptomatic infections [2]. The symptoms of COVID-19 vary from non-severe such as fever, cough, sore throat, malaise, headache, muscle pain, nausea, vomiting, diarrhea, loss of taste and smell to severe symptoms such as acute respiratory distress syndrome (ARDS) [3]. Severe COVID-19 cases and deaths mostly comprise patients with co-morbidities and immunocompromised diseases [4]. Moreover, a study conducted by Al-Awaida et al. (2021) on SARS-CoV-2 genome sequences from different continents for the period of 1 December 2020 to 15 March 2021 have showed a positive correlation of SARS-CoV-2 variant generation by mutations to number of cases and deaths per millions [5]. Fortunately, emergency use approval of the first COVID-19 vaccine, BNT162b2 (31 December 2020) followed by others such as AZD1222 (15 February 2021), Ad26-COV2.S (12 Mac 2021), mRNA-1273 (30 April 2021), BBIBP-CorV (7 May 2021) and CoronaVac (1 June 2021) has successfully reduced the incidence of severe COVID-19 [6]. However, COVID-19 vaccines are not preventing infections or reinfections by new SARS-CoV-2 variants [7, 8]. Current COVID-19 antiviral drugs such as remdesivir (Veklury), nirmatrelvir/ritonavir (Paxlovid) and molnupiravir (Lagevrio) were reported to be less effective in COVID-19 patients with comorbidities and immunocompromised diseases [9,10,11,12,13,14].
The different stages of COVID-19 pathogenesis such as early viral illness, inflammatory lung injury, and post-acute sequelae have caused finding a single treatment that can stop the progression of severe COVID-19 becomes challenging [15]. Therefore, continuous efforts in finding alternative COVID-19 treatment candidates possessing virus-directed (inhibition of viral essential components) and host-directed (anti-inflammatory or immunomodulatory activities) properties could be an effective strategy. The potential of herbs or medicinal plant products and their active compounds focusing on virus-directed and host-directed therapies for COVID-19 treatment has been highlighted in many studies [16,17,18].
The idea of targeting both virus replication and host mechanisms in treating COVID-19 is another approach to reduce viremia and stop the severe COVID-19 progression, which mainly caused by hyperinflammation or cytokine storm in the infected lungs [19]. Examples of potential agents possessing viral-directed and host directed activities are curcumin, a major compound of Curcuma longa L. and Huashi Baidu a traditional Chinese herbal mixture product. Curcumin was showed to inhibit replication of different SARS-CoV-2 strains potentially by targeting viral S protein-host ACE2 receptor interaction and inhibiting SARS-CoV-2 main protease enzyme (Mpro/3CL protease) [20,21,22,23]. A systematic review study on 6 curcumin clinical trials of hospitalized COVID-19 patients has suggested the potential immunomodulatory role of curcumin by restoring the pro-inflammatory and anti-inflammatory balance hence led to amelioration of cytokine storm manifestation [24]. A study conducted by Xu et al. (2023) on Huashi Baidu (Q-14), which consists of 14 herbs mixture, has discovered antiviral (magnolol, glycyrrhisoflavone, licoisoflavone A, emodin, echinatin and quercetin) and anti-inflammatory agents (licochalcone B, echinatin and also glycyrrhisoflavone) against SARS-CoV-2 infection in vitro [18]. In detail, echinatin and quercetin were potent inhibitor of SARS-CoV-2 Mpro or 3CL protease while glycyrrhisoflavone and licoisoflavone A were potent inhibitors of RNA-dependent RNA polymerase (RdRp). Glycyrrhisoflavone and licoisoflavone A also displayed potent inhibitory activities against cAMP-specific 3’, 5’-cyclic phosphodiesterase 4 (PDE4), an important enzyme in pro-inflammatory pathway [18].
In Malaysia, there is a medicinal plant that is worth exploring called Marantodes pumilum or also known as Labisia pumila [25, 26]. This plant, locally known as “Kacip fatimah”, is traditionally consumed by Malay women for induction and facilitation of labour, as well as to treat flatulence, dysentery, dysmenorrhoea, gonorrhoea and “sickness in the bones” [27]. Many scientific studies have been conducted to support the traditional claims of this plant. The antioxidant, phytoestrogenic and anti-inflammatory properties of M. pumilum have been shown to facilitate the prevention and treatment of diseases related to estrogen deficiencies [28]. The consumption of M. pumilum extracts showed beneficial effects in women and rat models with postmenopausal syndrome [29,30,31], polycystic ovary syndrome [32] and osteoporosis [33,34,35,36]. In addition, the beneficial effects of M. pumilum were also explored and studied for other diseases including diabetes [37, 38], cancer [25, 39] and bacterial infections [40, 41]. M. pumilum is rich in bioactive compounds such as catechins, rutin, quercetin, naringenin and gallic acids, which have been studied for anti-inflammatory and anti-SARS-CoV-2 activities in silico and in vitro [16, 17, 42,43,44,45,46,47,48,49,50]. However, scientific evidence to support the anti-COVID-19 potential of M. pumilum extract or product is still lacking.
In this study, our objectives are to evaluate the virus-directed therapy (anti-SARS-CoV-2 activity) and host-directed therapy (anti-inflammatory activity) potentials of M. pumilum standardized extract (SKF7®) in vitro. Anti-SARS-CoV-2 study of the SKF7® was tested against wild-type (Wuhan) and omicron SARS-CoV-2 variants. The effect of the SKF7® on viral S protein-human ACE2 interaction and viral 3CL protease or main protease (Mpro) activities were also evaluated for potential mechanism of action. Hyperinflammation is the key factor that contributed to the progression of severe COVID-19. Therefore, the SKF7® was also evaluated for potential anti-inflammatory effects against nitric oxide (NO) production and toll like receptor 2/4 (TLR2/4) responses, which were previously reported to be dysregulated during SARS-CoV-2 infections [51,52,53,54,55].
Methods
Test items
The SKF7®, a hydroalcoholic extract from a dried whole plant of M. pumilum (Blume) Kuntze was supplied by Medika Natura Sdn Bhd, a Malaysian biopharmaceutical company. The extract was standardized for not less than 2% gallic acid content (3.09%). Inhibitor control compounds for anti-SARS-CoV-2 (nirmatrelvir) and anti-inflammatory (curcumin) activities were purchased from TargetMol (TargetMol, USA). All test items were dissolved in 100% DMSO and kept at -20°C prior to use.
Tentative identification of SKF7® compounds by ultra-high performance liquid chromatography-high resolution accurate mass spectrometry (UHPLC-HRAMS)
The SKF7® extract was dissolved in water (5 mg/mL) and filtered through 0.22 μm PTFE filter prior to injection into the liquid chromatrography-mass spectrophotometry (LCMS) system. All solvents used for the preparation of the sample and mass spectrometry analysis was LCMS graded solvent purchased from Fisher Chemical (Geel, Belgium). The analysis was performed by using the Thermo Scientific Dionex Ultimate 3000 Series RS pump coupled with a Thermo Scientific Dionex Ultimate 3000 Series TCC-3000RS column compartments and a Thermo Fisher Scientific Ultimate 3000 Series WPS-3000RS autosampler controlled by Chromeleon 7.2 Software (Thermo Fisher Scientific, Waltham, MA and Dionex Softron GMbH Part of Thermo Fisher Scientific, Germany). Separations were performed by using an ACQUITY UPLC® BEH C18 analytical column (2.1 mm × 100 mm; particle size, 1.7 μm) (Waters, Milford, MA, USA) equipped with a Van Guard BEH C18 pre-column (2.1 mm × 5 mm; particle size, 1.7 m) (Waters, Milford, MA, USA) maintained at 40 °C. The mobile phase consisted of solutions A (0.1%v/v formic acid in water) and B (0.1% v/v formic acid in acetonitrile solution). A gradient program was used for elution including 5% solution B initially for 2 min, followed by 5–95% solution B from 2 to 15 min, 95% solution B from 15 to 18 min, and 5% solution B from 18.5 to 20 min. The mobile phase was delivered at a flow rate of 0.3 mL per min with an injection volume of 1 µL. HRAMS data were acquired using a Thermo Scientific Q-Exactive Orbitrap mass spectrometer controlled by the Xcalibur 3.0.63 software (Thermo Fisher Scientific, Waltham, MA) and operated at 70,000 resolution in full scan and 35,000 in MS/MS scan mode. The key heat electrospray ionization (HESI) source parameters were optimized as follows: spray voltage, 4.0 kV; capillary temperature, 320 °C, sheath gas flow rate 35; auxiliary gas flow rate 102; heater temp, 350 °C; S-lens RF level 55. The spectrum data type was centroid with a scan range from 100 to 1500 M/Z. The target value (AGC) was 1 × 105 and the maximum allowed accumulation time (IT) was 60 ms. For the data-dependent, MS/MS (ddMS2) analyses a TopN = 5 method was used. The five most intense peaks were selected for fragmentation with stepped normalized energy of 15, 30, and 45 V in negative ionization mode.
Cell culture and maintenance
African green monkey-derived epithelial kidney cells (Vero E6; ATCC-CRL-1586) and murine macrophage cells (RAW 264.7; ATCC TIB-71) were purchased from the American Type Cell Culture Collection (ATCC, USA). Human lung carcinoma expressing the human angiotensin I-converting enzyme-2 (hACE2) and transmembrane protease serine 2 (TMPRSS2) (A549-hACE2-TMPRSS2), and human toll-like receptor/NF-kB-SEAP reporter HEK293 (HEK-Blue hTLR2 and HEK-Blue hTLR4) cells were purchased from InvivoGen (InvivoGen, USA). Vero E6 and RAW 264.7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)(Gibco, USA) supplemented with 10% fetal bovine serum (FBS)(Gibco, USA) and 100 units/mL penicillin-streptomycin (Gibco, USA). The A549-hACE2-TMPRSS2 cell was cultured in DMEM (Gibco, USA) supplemented with 10% FBS (Gibco, USA), 100 µg/mL normocin (InvivoGen, USA), 100 units/mL penicillin-streptomycin (Gibco, USA), 0.5 µg/mL puromycin (InvivoGen, USA) and 300 µg/mL hygromycin (InvivoGen, USA). All cell lines were grown at 37°C in 5% CO2 incubator. HEK-Blue hTLR2 and HEK-Blue hTLR4 cells were cultured in DMEM (Gibco, USA) supplemented with 10% FBS (Gibco, USA), 100 µg/ml normocin (InvivoGen, USA), 100 units/mL penicillin-streptomycin (Gibco, USA) and 1X HEK-Blue Selection (InvivoGen, USA).
Virus propagation and titer determination
Clinical SARS-CoV-2 variants, Wuhan wild-type (WT) and Omicron (B.1.1.529) were previously isolated and cultured by the Virology Unit, Institute for Medical Research, National Institutes of Health, Malaysia. Virus isolation from human samples was not considered as human subject research and the requirement for written informed consent was exempted by the Medical Research and Ethics Committee (MREC), Ministry of Health, Malaysia. Virus isolates were propagated in Vero E6 cell culture containing 2% FBS supplemented DMEM media at 37°C in 5% CO2 incubator at biosafety level 3 (BSL-3) lab facility, Institute for Medical Research, National Institutes of Health, Malaysia. Once the viral induced cytopathic effect (CPE) was observed, the viral culture supernatant was collected and kept in -80°C prior to use.
The virus titer was measured by 50% tissue culture infective dose (TCID50) method. Vero-E6 (2 × 104 cells/well) and A549-hACE2-TMPRSS2 (1 × 104 cells/well) cells were seeded in 96-well culture plates and grown overnight. The cells were exposed to serially diluted (10-fold) viruses and incubated for 72 h at 37°C in 5% CO2 incubator. The CPE was observed microscopically. The viral induced CPE level was measured by ATP-based assay (ViralToxGlo®, Promega, USA). The luminescent signal intensity was measured by multimode microplate reader (GloMax® Explorer, Promega, USA). The TCID50 values were determined by dose-response curve analysis using GraphPad Prism software (Version 7.0).
Determination of anti-SARS-CoV-2 potential activity of the SKF7®
Cell viability assay
The nature of antiviral screening by CPE inhibition assay is by measuring cell death caused by virus infection. Therefore, the dose effect of the SKF7® on healthy cell viability need to be determined so that the non-cytotoxic concentrations are used in the antiviral screening. Briefly, Vero (2 × 104 cells/well) or A459-hACE2-TMPRSS2 cells (1 × 104 cells/well) were seeded in a 96-well culture plate containing DMEM media supplemented with 2% FBS and 100 units/mL penicillin-streptomycin (Gibco, USA) and incubated overnight at 37°C in 5% CO2 incubator. Then, confluent cells were exposed to serially diluted concentrations of the SKF7® (Vero: 3.13–800 µg/mL and A459-hACE2-TMPRSS2: 0.39–100 µg/mL) or nirmatrelvir (0.39–100 µM) and incubated for 72 h at 37°C in 5% CO2 incubator. The healthy cell control was cells exposed to media containing 0.5% DMSO only and the lysis cell control was cells exposed to media containing 0.05% Triton X 100. The cell viability level was measured by ATP-based assay. The ATP detection reagent (CellTiter-Glo®, Promega, USA) was added and incubated at room temperature for 10 min. The luminescent signal intensity was measured by multimode microplate reader (GloMax® Explorer, Promega, USA). The half-maximal cytotoxic concentration value (CC50) was determined by dose-response curve analysis using GraphPad Prism software (Version 7.0). The assay was performed in quadruplicate in three independent tests.
Antiviral screening by SARS-CoV-2 induced-cytopathic effect (CPE) inhibition assay
The Vero E6 cells were seeded at 2 × 104 cells/well in a 96 well plate and incubated overnight in DMEM media supplemented with 2% FBS and 100 units/mL penicillin-streptomycin (Gibco, USA) at 37°C in 5% CO2 incubator. Then, the serially diluted SKF7® (0.39–25 µg/mL) or SARS-CoV-2 inhibitor control, nirmatrelvir (0.39–100 µM) were added to the confluent cells. In the BSL3 lab, SARS-CoV-2 virus isolate was diluted with DMEM media supplemented with 2% FBS and added to the confluent cells in 96 well plate (25TCID50 per well) containing serially diluted extract or inhibitor control. The healthy cell control was cells exposed to media containing 0.5% DMSO only and the CPE control was cells exposed to the virus only. The test plate was incubated for 72 h at 37°C in 5% CO2 incubator. The viral cytopathic effect (CPE) was measured by ATP-based assay (ViralToxGlo®, Promega, USA). The luminescent signal intensity was measured by multimode microplate reader (GloMax® Explorer, Promega, USA). The half-maximal effective concentration value (EC50) was determined by dose-response curve analysis using GraphPad Prism software (Version 7.0). The assay was performed in quadruplicate in three independent tests.
Anti-SARS-CoV-2 screening by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
SARS-CoV-2 infection in human lung cells (A459-hACE2-TMPRSS2) did not induce measurable CPE within 72 h of incubation period. Therefore, RT-qPCR was performed to quantitate the viral RNA level affected by the SKF7® treatment. Briefly, A459-hACE2-TMPRSS2 cells were seeded (1 × 104 cells/well) in a 96 well culture plate containing DMEM media supplemented with 2% FBS, 100 µg/mL normocin (InvivoGen, USA), 100 units/mL penicillin-streptomycin (Gibco, USA), 0.5 µg/mL puromycin (InvivoGen, USA) and 300 µg/mL hygromycin (InvivoGen, USA), and incubated overnight at 37°C in 5% CO2 incubator. Then, the SKF7® extract (0.098–25 µg/mL), which was serially diluted with 2% FBS supplemented DMEM media, were added to the confluent cells. In the biosafety level 3 (BSL3) laboratory, SARS-CoV-2 virus isolate was diluted with 2% FBS supplemented DMEM media and added to the confluent cells in 96 well plate (0.03 MOI) containing serially diluted concentration of the SKF7® extract. The healthy cell control was cells exposed to media containing 0.5% DMSO only and the infection control was cells exposed to the virus only. The test plate was incubated for 72 h at 37°C in 5% CO2 incubator. The viral RNA was extracted from the culture supernatant by KingFischer Duo Prime System (ThermoFischer Scientific, USA) using a method described by the MagMax™ Viral/Pathogen Nucleic Acid Isolation Kit (Applied Biosystems™, USA). The primers targeting the N gene region [56] were used for the viral RNA copy number quantitation by adapting the RT-qPCR condition as recommended by Quantitect SYBR Green RT-PCR Kit (Qiagen, Germany). The RT-qPCR was performed by using the Applied Biosystems 7500 thermocycler (Applied Biosystems, USA). The synthetic SARS-CoV-2 RNA (VR-3276SD) (ATCC, USA) with known copy number was used as a standard for viral RNA copy number quantitation in this assay. The assay was performed in triplicate in three independent tests.
Determination of anti-SARS-CoV-2 specific action mechanisms
Spike S1 (SARS-CoV-2)-ACE2 interaction inhibitor screening
The effect of the SKF7® on the spike S1-ACE2 interaction was determined by using the Spike S1 receptor binding domain (RBD) (SARS-CoV-2): ACE2 inhibitor screening colorimetric assay kit (BPS Bioscience, Canada). Briefly, the extract (5, 10 and 20 µg/mL) was added to the spike S1, Fc fusion, avi-tag (2 µg/mL) pre-coated 96 well plate together with the reaction mixture containing 1x immunobuffer and ACE2-Biotin (0.4 ng/µL). The positive control was the reaction mixture without the extract. The reaction mixture was incubated for 10 min at room temperature prior to 1 h incubation with streptavidin-horse radish peroxidase (HRP). After 1 h incubation, the HRP substrate was added for 5 min incubation, and the reaction was stopped by adding 1 M sulfuric acid. The signal intensity was measured at 450 nm by using microplate reader ((FLUOstar Omega, BMG LABTECH, Germany). The assay was performed in triplicate.
3CL protease (SARS-CoV-2) assay
The effect of the SKF7® on 3CL protease or main protease (Mpro) activity was determined by using the 3CL protease, untagged (SARS-CoV-2) Assay kit (BPS Bioscience, Canada). Briefly, the extract (5, 10 and 20 µg/mL) was incubated with the reaction mixture containing assay buffer and 3CL protease (0.3 ng/µL). The positive control was the reaction mixture without the extract or inhibitor control. The inhibitor control was protease inhibitor GC376 (100 µM). The reaction mixture was preincubated for 20 min at room temperature with slow shaking. The reaction was started by adding 3CL protease substrate (40 µM) and incubated at room temperature for 4 h with slow shaking. The fluorescence intensity was measure at excitation wavelength 350 nm and emission wavelength 460 nm by using microplate reader (FLUOstar Omega, BMG LABTECH, Germany). The assay was performed in triplicate.
Determination of anti-inflammatory action mechanisms
Nitric oxide assay
The RAW 264.7 cells were seeded in 24-well plates at concentration 5 × 105 cells/well in 500 µL total volume with or without LPS (1 µg/mL) at 37 °C in a 5% CO2 incubator. After 4 h, the cells were treated with various concentrations of the SKF7® (1.56 to 100 µg/mL) for 24 h. Untreated-unstimulated cells and LPS-stimulated cells served as negative control and positive control, respectively. The inhibitor control compound was curcumin (12.5 µM). Nitrite level in the culture media, which represents intracellular nitric oxide synthase activity, were determined by Griess reaction. Briefly, cell supernatants were dispensed into 96-well plates and 150 µL of each supernatant was mixed with 20 µL of Griess reagent (Invitrogen, USA) and 130 µL of deionized water. The plate was then incubated at room temperature for 10 min. The concentration of NO was determined by a standard curve prepared with known concentrations of sodium nitrite as standard and the absorbance was measured at 550 nm by using plate reader (FLUOstar Omega, BMG LABTECH, Germany). The assay was performed in triplicate.
Toll-like receptor blocker assay
The HEK-Blue-hTLRs was produced by co-transfection of the human TLRs and CD14 co-receptor genes, and an inducible SEAP (secreted embryonic alkaline phosphatase) reporter gene into HEK293 cells (human embryo kidney cells) (InvivoGen, USA). The HEK-Blue hTLR2 and HEK-Blue hTLR4 cells were plated at the density of 1 × 105 cells/well in 96-well plates in pre-warmed HEK-Blue™ Detection Media (InvivoGen, USA). The LPS (1 µg/mL) and various concentrations of the SKF7® (0.78 to 100 µg/mL) were added to the cells and incubated overnight at 37°C in 5% CO2 incubator. The positive control was cells incubated with the LPS only while the negative control was cells without LPS induction. The inhibitor control compound was curcumin (12.5 µM). After 24 h of stimulation, the level of SEAP protein secretion, which represents the TLR activation, was quantified by using a spectrophotometer at 630 nm (FLUOstar Omega, BMG LABTECH, Germany). The assay was performed in triplicate in three independent tests.
Data analysis
The half-maximal effective (EC50) and cytotoxic (CC50) concentrations were generated from the dose-response curve analysis or nonlinear regression (curve fit) XY analysis using [inhibitor] versus normalized response (variable slope) algorithm in the GraphPad Prism software, version 7.0. The CC50 and EC50 values were used to calculate the selectivity index (SI) value (CC50/EC50). The significant changes (P < 0.05) between groups were analyzed by one-way ANOVA with Tukey’s multiple comparison analysis.
Results
The SKF7® is rich in phenolic compounds
The LCMS analysis was performed to identify the phytochemical content of the SKF7® extract. A total of 13 peaks were detected from the LCMS chromatogram (Figs. 1) and 9 of the peaks were tentatively identified as malic acid, quinic acid, gallic acid, gallocatechin, epigallocatechin, catechin, epigallocatechin gallate, ethyl gallate and epicatechin gallate (Additional file 1).
The LCMS chromatogram of the the SKF7® extract. Thirteen peaks consist of 9 tentatively identified phytochemical compounds and 4 unidentified compounds. The tentatively identified phytocompounds are (1) malic acid, (2) quinic acid, (3) gallic acid, (4) gallocatechin, (5) epigallocatechin, (6) catechin, 9) epigallactocatechin gallate, 10) ethyl gallate and 11) epicatechin gallate
The SKF7® inhibited SARS-CoV-2 infections in the kidney and lung cells
The cell viability assay or ATP-based assay was conducted to determine the optimum non-cytotoxic concentrations of the SKF7®, which will be used for the anti-SARS-CoV-2 testing. The result showed that the CC50 value of the SKF7® on Vero E6 cells was 158.50 µg/mL (Fig. 2). The highest starting concentration of the extract that showed more than 80% cell viability, which was 25 µg/mL, was used in the anti-SARS-CoV-2 testing (Fig. 2).
The cytotoxic effect of the SKF7® on the Vero E6 cells. The highest concentration of the extract that showed > 80% cell viability was at 25 µg/mL. The Vero E6 monolayer cells were exposed with serially diluted concentration of the SKF7® for 72 h at 37°C in 5% CO2 incubator. The cytotoxic activity was measured by CellTiter-Glo assay kit (Promega, USA). The plotted data are the mean value of cell viability percentage ± standard error of mean (SEM) from 3 independent tests
The anti-SARS-CoV-2 potential of the SKF7® was tested against wild-type (WT Wuhan) and omicron SARS-CoV-2 variants. The results showed that the SKF7® inhibited the WT Wuhan and the omicron SARS-CoV-2 infections (Fig. 3) as measured by the reduction in viral-induced CPE in Vero E6 cells. The EC50 values of the SKF7® against the WT Wuhan and the omicron SARS-CoV-2 infections were 21.99 µg/mL and 16.29 µg/mL, respectively (Fig. 3). The SI values for the SKF7® in inhibiting the WT Wuhan and the omicron SARS-CoV-2 infections in Vero E6 cells were 7.21 and 9.73, respectively (Fig. 3). The inhibitor control drug, nirmatrelvir showed potent antiviral activity against the WT Wuhan SARS-CoV-2 with an EC50 value of 1.26 µM and SI value of > 79.37 (Additional file 2).
The antiviral activity of the SKF7® against the WT Wuhan and the Omicron SARS-CoV-2 variants infections in the kidney cells. The Vero E6 monolayer cells incubated with various concentrations of the extract and the SARS-COV-2 virus (25TCID50) for 72 h at 37°C in 5% CO2 incubator. The percentage on infection was determined by the viral-induced CPE level as measured by ATP-based assay. The luminescent signal of ATP levels were normalized to the percentage of infection values. The plotted data are the mean value of infection percentage ± standard error of mean (SEM) from 3 independent tests
In order to recapitulate the antiviral activity in human lung infections, the anti-SARS-CoV-2 activity of the SKF7® was also evaluated in the human lung cells (A459-hACE2-TMPRSS2), which expressed the human ACE2 and TMPRSS2 receptors, infected with WT Wuhan SARS-CoV-2 variant. At the end of the incubation, the culture supernatant containing the mature virus was collected for viral RNA detection by RT-qPCR. The CPE based assay was not implemented in this experiment as the infected A549 cells were not showing measurable CPE within the 72 h of incubation period. The RT-qPCR results showed that the SKF7® reduced the SARS-CoV-2 infection in the A459-hACE2-TMPRSS2 cells with the EC50 value of 6.31 µg/mL (Fig. 4). The cytotoxic effect of the SKF7® against the A459-hACE2-TMPRSS2 cells was also determined by ATP-based assay and the CC50 value was 76.83 µg/mL (Fig. 4). This has resulted higher SI value (SI > 10) for the SKF7® against the WT Wuhan SARS-CoV-2 infections in A459-hACE2-TMPRSS2 cells (Fig. 4) as compared to WT Wuhan SARS-CoV-2 infections in Vero E6 cells (Fig. 3).
The SKF7® inhibited the WT Wuhan SARS-CoV-2 infections in human lung cells. The A459-hACE2-TMPRSS2 monolayer cells were incubated with various concentrations of the extract together with the SARS-COV-2 virus (MOI: 0.03) for 72 h at 37°C in 5% CO2 incubator. The viral RNA level from the culture supernatant was quantitated by RT-qPCR. The cell viability of the cells exposed with various concentration of the extract was measured by ATP-based assay. The viral RNA and cytotoxic levels were normalized to percentage of infection and cell viability, respectively. The plotted data are the mean value of infection or cell viability percentage ± standard error of mean (SEM) from 3 independent tests
The SKF7® prominently inhibited the SARS-CoV-2 3CL protease activity
The potential of the SKF7® to affect the essential enzymes involved in SARS-CoV-2 virus internalization and replication were determined by the spike S1-ACE2 and 3CL protease inhibitor assays, respectively. Our data showed that the spike S1-ACE2 interaction activity was not affected by the lower SKF7® concentrations (5 µg/mL and 10 µg/mL) (Fig. 5A). However, at higher concentration of the SKF7®, 20 µg/mL, the 13% reduction of the spike S1-ACE2 interaction activity was observed but it was not statistically significant (Fig. 5A).
Interestingly, the effect of the SKF7® was more prominent against the 3CL protease or main protease (Mpro) activity (Fig. 5B). The SKF7® significantly (P < 0.05) reduced the 3CL protease activity by 32.8% and 53.5% at the concentration of 10 µg/mL and 20 µg/mL, respectively (Fig. 5B). The insignificant reduction of 4.3% was observed at 5 µg/mL of the SKF7® (Fig. 5B). The control inhibitor, GC376 (100 µM), showed 100% reduction of the 3CL protease activity (Fig. 5B).
The effect of the SKF7® on the S1-ACE2 interaction and 3CL protease activities. The bar charts represent normalized mean percentages of S1-ACE interaction (A) and 3CL (B) activities (as compared to untreated control) ± standard deviation (SD) from 3 technical replicates. The asterisk (*) denotes a significant change (P < 0.05) as compared to untreated control group
The SKF7® inhibited the LPS-induced TLR4 activation
Besides the antiviral activity, the anti-inflammatory potential of the SKF7® was also evaluated. The general nitric oxide assay was performed on the LPS-induced murine macrophage cells (RAW 264.7) exposed with various concentrations (1.56–100 µg/mL) of the SKF7®. However, the SKF7® treatment did not affects the NO production in LPS-induced murine macrophage cells and the decrease in NO level was influenced by the reduction of cell viability due to dose-dependent cytotoxic effect of the extract. Dose-response curve analysis showed that the EC50 value of the SKF7® in reducing the NO level (67.68 µg/mL) was higher than the CC50 value (42.76 µg/mL). This resulted in the SI value of less than 1 (0.63) (Additional file 3).
The potential of the SKF7® in affecting TLR2 and TLR4 inflammatory signaling responses were also evaluated. The LPS-induced TLR2 signaling activation was not affected by the SKF7® treatment and the observed reduction of TLR2 signaling activity was in parallel with the reduction of HEK-Blue hTLR2 monolayer cell viability (Fig. 6A). This was further supported by the dose-response curve analysis where the EC50 value of the SKF7® in reducing the TLR2 activation was higher than the CC50 value. This resulted the SI value of less than 1 (< 1.07) (Fig. 6B).
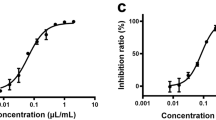
On the other hand, the SKF7® treatment inhibited the LPS-induced TLR4 signaling response in a dose dependent manner (Fig. 6C). As compared to the LPS induction alone, the TLR4 response was significantly inhibited (P < 0.05) by the SKF7® at the concentration of 25 µg/mL and 50 µg/mL (Fig. 6C). The TLR4 inhibition was not in the range of the SKF7® cytotoxic concentrations (Fig. 6C). As determined by dose response curve analysis, the EC50 and CC50 values of the SKF7® was 16.19 µg/mL and 67.30 µg/mL, respectively and the SI value was 4.16 (Fig. 6D). The curcumin, a positive control compound, showed > 80% inhibition of TLR2/4 signaling responses after LPS induction.
Effect of SKF7® treatment on the TLR2 and TLR4 signaling activations. HEK-Blue hTLR2 (A) or hTLR4 (C) monolayer cells were exposed with LPS (1 µg/mL) and various concentrations of the extract for 24 h at 37°C in 5% CO2 incubator. The level of secreted alkaline phosphatase protein (SEAP), which representing the TLRs activation, was quantified by using a spectrophotometer at 630 nm. Cytotoxic effects of the SKF7® on HEK-Blue hTLR2 and hTLR4 cells were measured by the ATP-based assay. The dose-response curve analysis of the SKF7® against the TLR2 (B) and TLR4 (D) was conducted by using the GraphPad prism software. Curcumin was used as an inhibitor control compound that inhibit both TLR2 and TLR4 activations. The charts represent normalized mean values of TLRs activation (%) or cell viability (%) ± standard deviation (SD) from 3 independent tests. The asterisk (*) denotes a significant change (P < 0.05) as compared to LPS-induced TLR2/4 activation alone group
Discussion
The potential of COVID-19 therapeutic candidates could be assessed through their direct antiviral and host directed activities. These activities include the ability of the candidate(s) to reduce viremia by inhibiting the SARS-CoV-2 entry and replication inside the host and to regulates the excessive inflammatory responses by reducing the pro-inflammatory molecules or inhibiting the signaling activity that causes the severity in COVID-19 patients [57]. During the COVID-19 pandemic (2020–2021), many medicinal plant products and compounds were pre-clinically studied for their potentials in COVID-19 treatments through direct antiviral therapy and/or host directed therapy [16, 20, 58,59,60].
To our knowledge, this is the first study to demonstrate the antiviral potential of M. pumilum standardized extract, the SKF7®, against SARS-CoV-2 infections in kidney (Vero) and lung (A459-hACE2-TMPRSS2) cells. In year 2020 to 2021, researchers in Thailand have highlighted two potential anti-COVID-19 medicinal plant candidates, Andrographis paniculata or “King of bitters” [59] and Boesenbergia rotunda or locally known as fingerroot [58]. Both plant extracts, A. paniculata and B. rotunda exhibited potent anti-SARS-CoV-2 in Vero E6 cells with EC50 value of 68.06 µg/mL (SI: >1.47) and 3.62 µg/mL (SI: 7.75), respectively [58]. The active compounds for A. paniculata (andrographolide) and B. rotunda (panduratin A) were demonstrated to specifically inhibit the main protease (Mpro) or 3CL protease, which is essential for SARS-CoV-2 virus replication [60,61,62]. As a comparison, the SKF7® showed moderate antiviral activity in Vero E6 cells (EC50: 16.29–21.99 µg/mL). Interestingly, in human lung cells (A459-hACE2-TMPRSS2), the SKF7® showed enhanced antiviral activity with an EC50 value of < 10 µg/mL (6.31 µg/mL) and SI value of > 10. Similar finding was also reported in a study of A. paniculata extract against SARS-CoV-2 infection in human lung cells (Calu3), which scored smaller EC50 value (9.54 µg/mL) [59] as compared to its antiviral activity in Vero E6 cells. The deviation of antiviral response in different cells could be due to high efflux activity in kidney cells such as Vero E6 [63].
The SKF7® may be targeting the viral replication mechanism as it prominently inhibited the 3CL protease activity (Mpro), an essential SARS-CoV-2 protease for virus replication. The 3CL protease is a valid anti-SARS-CoV-2 drug target because one of the COVID-19 antivirals, nirmatrelvir (active component of Paxlovid) was developed based on this target [64]. The anti-SARS-CoV-2 activity of the SKF7® could be due to the interactions of its phytochemical compounds. As detected by UHPLC-HRAMS, the SKF7® extract contains numerous phenolic compounds, which were previously studied for anti-SARS-CoV-2 activities in silico and in vitro. These compounds are catechins (catechin, epigallocatechin and epigallocatechin-3-gallate) and gallic acid. The epigallocatechin-3-gallate (EGCG), an active compound found in green tea, was previously demonstrated to inhibits SARS-CoV-2, MERS-CoV and SARS-CoV infections in Vero cells with the EC50 values of 1.73 µg/mL, 4.64 µg/mL and 0.83 µg/mL, respectively [46]. Indeed, an in-silico study has showed that out of all catechins, EGCG formed the best affinity and stable complex with the spike protein [47]. Another in silico study has also shown that EGCG together with other catechins (epicatechingallate and gallocatechine-3-galate) formed the best affinity and stable complex with the 3CL protease or Mpro [48]. Besides green tea, EGCG was also an active ingredient of Traditional Chinese Medicines (Yangyinjiedu, Dayuanxiaodu and Chaihuqingzao), which have been used for COVID-19 treatment in the hospitals in Guizhou Province, China [49]. A detailed in silico and in vitro analysis have showed that EGCG has higher binding affinity (-7.9 kcal/mol) towards the 3CL protease with a promising enzyme inhibitory activity (IC50 value of 0.847 µM) [49]. The SKF7® was standardized to not less than 2% gallic acid content (3.09%) in the native extract. A study conducted by Gu et al. (2023) showed that gallic acid bound tightly (Kd: 15 µM) to non-structural protein-7 (nsp7), one of essential components for SARS-CoV-2 replication [65]. However, our previous study has showed that gallic acid was not effective in inhibiting the SARS-CoV-2 infection in Vero E6 cells [20].
Apart from anti-SARS-CoV-2 activity, the SKF7® also possessed anti-inflammatory property as it inhibited the in vitro activity of LPS-induced TLR4 response, which was previously found to be one of the important pro-inflammatory signaling pathways that contributed to the COVID-19 pathogenesis during SARS-CoV-2 infections [51]. The actual compound that blocked the TLR4 response but not TLR2 remains to be elucidated. However, one of the potential compounds could be the EGCG, which was previously reported to affect the TLR4 signaling in various disease models in vitro and in vivo [66,67,68,69,70]. Other studies on M. pumilum extracts have highlighted the anti-inflammatory effects (by reducing the pro-inflammatory cytokines) in the postmenopausal osteoporosis, diabetes, and gout studies [71,72,73,74] but none of the studies emphasized on the TLRs blocker effect of the extracts. Theoretically, the pro-inflammatory cytokine production was governed by the toll like receptor (TLR) signaling pathways [75] through the interaction of cell surface TLRs with the membrane components of microorganism (lipids, lipoproteins and proteins) [75]. TLR4 is mainly expressed on immune cells (macrophages, dendritic cells, and monocytes) which are mostly resided in the lungs and heart [51]. Instead of its role in hyperinflammation induction SARS-CoV-2 infection [52, 55], the TLR4 was found to be a mediator in cognitive dysfunction induced by SARS-CoV-2 spike protein in mice, which recapitulating the post-COVID-19 syndrome [76]. Therefore, the potential of the SKF7® in inhibiting the TLR4 signaling response might contribute in preventing the progression of severe COVID-19 and treating the post-COVID-19 syndrome. However, this hypothesis needs to be confirmed by future in vivo studies.
As a discovery phase study, our in vitro data need to be supported by future pre-clinical experiments particularly an efficacy study in COVID-19 in vivo mouse model, which could highlights in detail on the effect of the SKF7® treatment on the COVID-19 pathogenesis such as viremia and inflammation of the target organ such as lung. Although 3CL protease inhibition activity was observed for the SKF7® antiviral mechanism, we could not exclude other potential mechanisms that maybe affected such as papain like protease (PLpro), which was not tested in the present study. The trend of infection inhibition by the SKF7® was limited to WT Wuhan and Omicron SARS-CoV-2 variants. Although the inhibition of 3CL protease could lead to a promising broad anti-SARS-COV-2 candidate as previously showed by nirmatrelvir [63], it still needs to be validated against other SARS-CoV-2 variants such as Delta and other Omicron sub-variants. The NO production in LPS-induced mouse macrophage cells (RAW 264.7) was not affected by the SKF7® treatment. It could be that the SKF7® and its compound(s) majorly targeting the TLR4 signaling pathway. However, the identity of the compound(s) with TLR4 blocking activity needs to be elucidated in future studies.
Conclusion
In conclusion, M. pumilum standardized extract (SKF7®), which was rich in phenolic compounds, possessed an anti-SARS-CoV-2 activity possibly by inhibiting the 3CL protease or Mpro activity. The SKF7® also inhibited the LPS-induced TLR4 activation activity in vitro, which indicates the potential anti-inflammatory role of this extract. Future pre-clinical in vivo studies focusing on antiviral and immunomodulatory potentials of the SKF7® in affecting the COVID-19 pathogenesis are warranted.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ATP:
-
Adenosine triphosphate
- BSL3:
-
Biosafety level 3
- CC50 :
-
Half maximal cytotoxic concentration
- COVID-19:
-
Coronavirus disease 2019
- CPE:
-
Cytopathic effect
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- DMSO:
-
Dimethylsulfoxide
- EC50 :
-
Half maximal effective concentration
- EGCG:
-
Epigallocatechin-3-gallate
- FBS:
-
Fetal bovine serum
- LPS:
-
Lipopolysaccharide
- MPro:
-
Main protease
- NO:
-
Nitric oxide
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- SI:
-
Selectivity index
- TCID50 :
-
Median tissue culture infectious dose
- TLR:
-
Toll-like receptor
- TMPRSS2:
-
Transmembrane serine protease 2
References
WHO. WHO Coronavirus (COVID-19.) Dashboard 2024 [updated 5 May 2024. https://covid19.who.int
Mugglestone MA, Ratnaraja NV, Bak A, Islam J, Wilson JA, Bostock J, et al. Presymptomatic, asymptomatic and post-symptomatic transmission of SARS-CoV-2: joint British Infection Association (BIA), Healthcare Infection Society (HIS), Infection Prevention Society (IPS) and Royal College of Pathologists (RCPath) guidance. BMC Infect Dis. 2022;22(1):453.
NIH. Clinical Spectrum of SARS-CoV-2 Infection 2023 [updated 6 March 2023. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/
Zhang JJ, Dong X, Liu GH, Gao YD. Risk and protective factors for COVID-19 morbidity, severity, and Mortality. Clin Rev Allergy Immunol. 2023;64(1):90–107.
Al-Awaida J, Jawabrah Al Hourani W, Swedan B, Nimer S, Alzoughool R, Al-Ameer FJ. H, Correlates of SARS-CoV-2 variants on deaths, Case Incidence and Case Fatality ratio among the continents for the period of 1 December 2020 to 15 March 2021. Genes [Internet]. 2021; 12(7).
Li M, Wang H, Tian L, Pang Z, Yang Q, Huang T, et al. COVID-19 vaccine development: milestones, lessons and prospects. Signal Transduct Target Therapy. 2022;7(1):146.
Chemaitelly H, Tang P, Hasan MR, AlMukdad S, Yassine HM, Benslimane FM, et al. Waning of BNT162b2 Vaccine Protection against SARS-CoV-2 infection in Qatar. N Engl J Med. 2021;385(24):e83.
Feikin DR, Higdon MM, Abu-Raddad LJ, Andrews N, Araos R, Goldberg Y, et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet. 2022;399(10328):924–44.
Liu J, Pan X, Zhang S, Li M, Ma K, Fan C et al. Efficacy and safety of Paxlovid in severe adult patients with SARS-Cov-2 infection: a multicenter randomized controlled study. Lancet Reg Health West Pac. 2023:100694.
Sun F, Lin Y, Wang X, Gao Y, Ye S. Paxlovid in patients who are immunocompromised and hospitalised with SARS-CoV-2 infection. Lancet Infect Dis. 2022;22(9):1279.
Del Borgo C, Garattini S, Bortignon C, Carraro A, Di Trento D, Gasperin A et al. Effectiveness, tolerability and prescribing choice of antiviral molecules Molnupiravir, Remdesivir and Nirmatrelvir/r: a real-world comparison in the First Ten months of Use. Viruses. 2023;15(4).
Tiseo G, Barbieri C, Galfo V, Occhineri S, Matucci T, Almerigogna F, et al. Efficacy and Safety of Nirmatrelvir/Ritonavir, Molnupiravir, and Remdesivir in a real-world cohort of outpatients with COVID-19 at high risk of progression: the PISA Outpatient Clinic Experience. Infect Dis Ther. 2023;12(1):257–71.
Malin JJ, Weibel S, Gruell H, Kreuzberger N, Stegemann M, Skoetz N. Efficacy and safety of molnupiravir for the treatment of SARS-CoV-2 infection: a systematic review and meta-analysis. J Antimicrob Chemother. 2023.
Butler CC, Hobbs FDR, Gbinigie OA, Rahman NM, Hayward G, Richards DB, et al. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet. 2023;401(10373):281–93.
Russell CD, Lone NI, Baillie JK. Comorbidities, multimorbidity and COVID-19. Nat Med. 2023;29(2):334–43.
Soleymani S, Naghizadeh A, Karimi M, Zarei A, Mardi R, Kordafshari G, et al. COVID-19: general strategies for Herbal therapies. J Evid Based Integr Med. 2022;27:2515690X211053641.
Jantan I, Norahmad NA, Yuandani, Haque MA, Mohamed-Hussein ZA, Mohd Abd Razak MR et al. Inhibitory effect of food-functioned phytochemicals on dysregulated inflammatory pathways triggered by SARS-CoV-2: a mechanistic review. Crit Rev Food Sci Nutr. 2024:1–26.
Xu H, Li S, Liu J, Cheng J, Kang L, Li W, et al. Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19. Proc Natl Acad Sci U S A. 2023;120(18):e2301775120.
Wallis RS, O’Garra A, Sher A, Wack A. Host-directed immunotherapy of viral and bacterial infections: past, present and future. Nat Rev Immunol. 2023;23(2):121–33.
Mohd Abd Razak MR, Md Jelas NH, Muhammad A, Padlan N, Sa’at MNF, Azizan MA, et al. In Vitro Anti-SARS-CoV-2 activities of Curcumin and selected phenolic compounds. Nat Prod Commun. 2023;18(9):1934578X231188861.
Marin-Palma D, Tabares-Guevara JH, Zapata-Cardona MI, Florez-Alvarez L, Yepes LM, Rugeles MT et al. Curcumin inhibits in Vitro SARS-CoV-2 infection in Vero E6 cells through multiple antiviral mechanisms. Molecules. 2021;26(22).
Bormann M, Alt M, van de Schipper L, Le-Trilling VTK, Rink L et al. Turmeric Root and its bioactive ingredient Curcumin effectively neutralize SARS-CoV-2 in Vitro. Viruses. 2021;13(10).
Jena AB, Kanungo N, Nayak V, Chainy GBN, Dandapat J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: insights from computational studies. Sci Rep. 2021;11(1):2043.
Vahedian-Azimi A, Abbasifard M, Rahimi-Bashar F, Guest PC, Majeed M, Mohammadi A et al. Effectiveness of Curcumin on outcomes of hospitalized COVID-19 patients: a systematic review of clinical trials. Nutrients. 2022;14(2).
Hanafi MMM, Yaakob H, Gibbons S, Prieto JM. In Vitro Pro-Apoptotic and Anti-Migratory Effects of Marantodes pumilum (syn. Labisia pumila) Extracts on Human Prostate Cancer Cell Lines: Bioguided Isolation of 5-Henicosene-1-yl-resorcinol. Plants (Basel). 2023;12(7).
Ibrahim IS, Mohd Said M, Mohammad Zainoor N, Jamal JA. Authentication of Marantodes pumilum (Blume) Kuntze: A Systematic Review. Front Pharmacol. 2022;13:855384.
Burkill IH. A Dictionary of the Economic Products of the Malay Peninsula. Kuala Lumpur, Malaysia: Ministry of Agriculture Malaysia; 1966.
Zakaria AA, Noor MHM, Ahmad H, Hassim HA, Mazlan M, Latip MQA. A Review on Therapeutic Effects of Labisia pumila on Female Reproductive Diseases. Biomed Res Int. 2021;2021:9928199.
Norhayati MN, George A, Hazlina NH, Azidah AK, Idiana HI, Law KS, et al. Efficacy and safety of Labisia pumila var alata water extract among pre- and postmenopausal women. J Med Food. 2014;17(8):929–38.
Abdul Kadir A, Nik Hussain NH, Wan Bebakar WM, Mohd DM, Wan Mohammad WM, Hassan II, et al. The Effect of Labisia pumila var. alata on Postmenopausal Women: A Pilot Study. Evid Based Complement Alternat Med. 2012;2012:216525.
Chinnappan SM, George A, Evans M, Anthony J. Efficacy of Labisia pumila and Eurycoma longifolia standardised extracts on hot flushes, quality of life, hormone and lipid profile of peri-menopausal and menopausal women: a randomised, placebo-controlled study. Food Nutr Res. 2020;64.
Manneras L, Fazliana M, Wan Nazaimoon WM, Lonn M, Gu HF, Ostenson CG, et al. Beneficial metabolic effects of the Malaysian herb Labisia pumila var. alata in a rat model of polycystic ovary syndrome. J Ethnopharmacol. 2010;127(2):346–51.
Mohd Effendy N, Abdullah S, Yunoh MF, Shuid AN. Time and dose-dependent effects of Labisia pumila on the bone strength of postmenopausal osteoporosis rat model. BMC Complement Altern Med. 2015;15:58.
Effendy NM, Shuid AN. Time and dose-dependent effects of Labisia pumila on bone oxidative status of postmenopausal osteoporosis rat model. Nutrients. 2014;6(8):3288–302.
Fathilah SN, Mohamed N, Muhammad N, Mohamed IN, Soelaiman IN, Shuid AN. Labisia pumila regulates bone-related genes expressions in postmenopausal osteoporosis model. BMC Complement Altern Med. 2013;13:217.
Fathilah SN, Abdullah S, Mohamed N, Shuid AN. Labisia pumila Prevents Complications of Osteoporosis by Increasing Bone Strength in a Rat Model of Postmenopausal Osteoporosis. Evid Based Complement Alternat Med. 2012;2012:948080.
Nahar N, Mohamed S, Mustapha NM, Fong LS. Protective effects of Labisia pumila against neuropathy in a diabetic rat model. J Diabetes Metab Disord. 2022;21(1):1–11.
Nahar N, Mohamed S, Mustapha NM, Fong LS, Mohd Ishak NI. Gallic acid and myricetin-rich Labisia pumila extract mitigated multiple diabetic eye disorders in rats. J Food Biochem. 2021;45(11):e13948.
Zakaria N, Mohd KS, Ahmed Saeed MA, Ahmed Hassan LE, Shafaei A, Al-Suede FSR, et al. Anti-Uterine Fibroid Effect of Standardized Labisia Pumila Var. Alata Extracts In Vitro and in Human Uterine Fibroid Cancer Xenograft Model. Asian Pac J Cancer Prev. 2020;21(4):943–51.
Fazliana M, Ramos NL, Luthje P, Sekikubo M, Holm A, Wan Nazaimoon WM, et al. Labisia pumila var. alata reduces bacterial load by inducing uroepithelial cell apoptosis. J Ethnopharmacol. 2011;136(1):111–6.
Karimi E, Jaafar HZ, Ghasemzadeh A, Ebrahimi M. Fatty acid composition, antioxidant and antibacterial properties of the microwave aqueous extract of three varieties of Labisia pumila Benth. Biol Res. 2015;48(1):9.
Sancineto L, Ostacolo C, Ortega-Alarcon D, Jimenez-Alesanco A, Ceballos-Laita L, Vega S et al. l-Arginine Improves Solubility and ANTI SARS-CoV-2 Mpro Activity of Rutin but Not the Antiviral Activity in Cells. Molecules. 2021;26(19).
El Gizawy HA, Boshra SA, Mostafa A, Mahmoud SH, Ismail MI, Alsfouk AA, et al. Pimenta dioica (L.) Merr. Bioactive Constituents Exert Anti-SARS-CoV-2 and Anti-Inflammatory Activities: Molecular Docking and Dynamics, In Vitro, and In Vivo Studies. Molecules. 2021;26:19.
Bahun M, Jukic M, Oblak D, Kranjc L, Bajc G, Butala M, et al. Inhibition of the SARS-CoV-2 3CL(pro) main protease by plant polyphenols. Food Chem. 2022;373Pt B:131594.
Teli DM, Shah MB, Chhabria MT. In silico Screening of Natural Compounds as Potential Inhibitors of SARS-CoV-2 Main Protease and Spike RBD: Targets for COVID-19. Front Mol Biosci. 2020;7:599079.
Henss L, Auste A, Schurmann C, Schmidt C, von Rhein C, Muhlebach MD et al. The green tea catechin epigallocatechin gallate inhibits SARS-CoV-2 infection. J Gen Virol. 2021;102(4).
Mhatre S, Gurav N, Shah M, Patravale V. Entry-inhibitory role of catechins against SARS-CoV-2 and its UK variant. Comput Biol Med. 2021;135:104560.
Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. 2021;39(12):4362–74.
Du A, Zheng R, Disoma C, Li S, Chen Z, Li S, et al. Epigallocatechin-3-gallate, an active ingredient of Traditional Chinese Medicines, inhibits the 3CLpro activity of SARS-CoV-2. Int J Biol Macromol. 2021;176:1–12.
Clementi N, Scagnolari C, D’Amore A, Palombi F, Criscuolo E, Frasca F, et al. Naringenin is a powerful inhibitor of SARS-CoV-2 infection in vitro. Pharmacol Res. 2021;163:105255.
Aboudounya MM, Heads RJ. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators Inflamm. 2021;2021:8874339.
Zhao Y, Kuang M, Li J, Zhu L, Jia Z, Guo X, et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021;31(7):818–20.
Khan S, Shafiei MS, Longoria C, Schoggins JW, Savani RC, Zaki H. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife. 2021;10.
Zheng M, Karki R, Williams EP, Yang D, Fitzpatrick E, Vogel P, et al. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat Immunol. 2021;22(7):829–38.
Shirato K, Kizaki T. SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon. 2021;7(2):e06187.
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–68.
Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–54.
Kanjanasirirat P, Suksatu A, Manopwisedjaroen S, Munyoo B, Tuchinda P, Jearawuttanakul K, et al. High-content screening of Thai medicinal plants reveals Boesenbergia rotunda extract and its component Panduratin A as anti-SARS-CoV-2 agents. Sci Rep. 2020;10(1):19963.
Sa-Ngiamsuntorn K, Suksatu A, Pewkliang Y, Thongsri P, Kanjanasirirat P, Manopwisedjaroen S, et al. Anti-SARS-CoV-2 Activity of Andrographis paniculata Extract and Its Major Component Andrographolide in Human Lung Epithelial Cells and Cytotoxicity Evaluation in Major Organ Cell Representatives. J Nat Prod. 2021;84(4):1261–70.
Lim XY, Chan JSW, Tan TYC, Teh BP, Mohd Abd Razak MR, Mohamad S, et al. Andrographis paniculata (Burm. F.) Wall. Ex Nees, Andrographolide, and Andrographolide Analogues as SARS-CoV-2 Antivirals? A Rapid Review. Nat Prod Commun. 2021;16(5):1934578X211016610.
Enmozhi SK, Raja K, Sebastine I, Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn. 2021;39(9):3092–8.
Vergoten G, Bailly C. Interaction of panduratin A and derivatives with the SARS-CoV-2 main protease (m(pro)): a molecular docking study. J Biomol Struct Dyn. 2022:1–11.
Vangeel L, Chiu W, De Jonghe S, Maes P, Slechten B, Raymenants J, et al. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res. 2022;198:105252.
Drozdzal S, Rosik J, Lechowicz K, Machaj F, Szostak B, Przybycinski J, et al. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist Updat. 2021;59:100794.
Gu Y, Liu M, Staker BL, Buchko GW, Quinn RJ. Drug-Repurposing Screening Identifies a Gallic Acid Binding Site on SARS-CoV-2 Non-structural Protein 7. ACS Pharmacol Transl Sci. 2023;6(4):578–86.
Hou H, Yang W, Bao S, Cao Y. Epigallocatechin Gallate Suppresses Inflammatory Responses by Inhibiting Toll-like Receptor 4 Signaling and Alleviates Insulin Resistance in the Livers of High-fat-diet Rats. J Oleo Sci. 2020;69(5):479–86.
Zhong X, Liu M, Yao W, Du K, He M, Jin X, et al. Epigallocatechin-3-Gallate Attenuates Microglial Inflammation and Neurotoxicity by Suppressing the Activation of Canonical and Noncanonical Inflammasome via TLR4/NF-kappaB Pathway. Mol Nutr Food Res. 2019;63(21):e1801230.
Shen H, Wu N, Liu Z, Zhao H, Zhao M. Epigallocatechin-3-gallate alleviates paraquat-induced acute lung injury and inhibits upregulation of toll-like receptors. Life Sci. 2017;170:25–32.
Xu MJ, Liu BJ, Wang CL, Wang GH, Tian Y, Wang SH, et al. Epigallocatechin-3-gallate inhibits TLR4 signaling through the 67-kDa laminin receptor and effectively alleviates acute lung injury induced by H9N2 swine influenza virus. Int Immunopharmacol. 2017;52:24–33.
Bao S, Cao Y, Zhou H, Sun X, Shan Z, Teng W. Epigallocatechin gallate (EGCG) suppresses lipopolysaccharide-induced Toll-like receptor 4 (TLR4) activity via 67 kDa laminin receptor (67LR) in 3T3-L1 adipocytes. J Agric Food Chem. 2015;63(10):2811–9.
Rahmi EP, Kumolosasi E, Jalil J, Husain K, Buang F, Abd Razak AF, et al. Anti-hyperuricemic and Anti-inflammatory Effects of Marantodes pumilum as Potential Treatment for Gout. Front Pharmacol. 2020;11:289.
Nadia ME, Nazrun AS, Norazlina M, Isa NM, Norliza M, Ima Nirwana S. The Anti-Inflammatory, Phytoestrogenic, and Antioxidative Role of Labisia pumila in Prevention of Postmenopausal Osteoporosis. Adv Pharmacol Sci. 2012;2012:706905.
Karim K, Giribabu N, Salleh N. Marantodes pumilum Var Alata (Kacip Fatimah) ameliorates derangement in RANK/RANKL/OPG pathway and reduces inflammation and oxidative stress in the bone of estrogen-deficient female rats with type-2 diabetes. Phytomedicine. 2021;91:153677.
Rahmi EP, Jamal JA, Kumolosasi E, Jalil J, Aladdin NA. Marantodes pumilum (Blume) Kuntze Inhibited Secretion of Lipopolysaccharide- and Monosodium Urate Crystal-stimulated Cytokines and Plasma Prostaglandin E(2). Pharmacogn Mag. 2017;13(Suppl 3):S578–86.
Duan T, Du Y, Xing C, Wang HY, Wang RF. Toll-Like Receptor Signaling and Its Role in Cell-Mediated Immunity. Front Immunol. 2022;13:812774.
Fontes-Dantas FL, Fernandes GG, Gutman EG, De Lima EV, Antonio LS, Hammerle MB, et al. SARS-CoV-2 Spike protein induces TLR4-mediated long-term cognitive dysfunction recapitulating post-COVID-19 syndrome in mice. Cell Rep. 2023;42(3):112189.
Acknowledgements
We would like to thank the Director General of Health Malaysia for his permission to publish this article. We also would like to thank the staff of Bioassay and Virology Units for their supports and contributions in this study.
Funding
This study was funded by Ministry of Health Malaysia [NMRR-20-874-54715].
Author information
Authors and Affiliations
Contributions
Conceptualization, MRMAR, MZ, and AFSM; Formal analysis, MRMAR and NAN; Funding acquisition, MRMAR; Methodology, MRMAR; Antiviral assays, MRMAR, AM, MNFS and NHMJ; Anti-inflammatory assays, NAN; Phytochemistry analysis, NMM; Project administration, MRMAR; Supervision, MZ and AFSM; Writing – original draft, MRMAR; Writing – review & editing, MRMAR, NAN, NHMJ, MZ and AFSM. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study has been given an expedited ethical approval including a waiver for informed consent by the Medical Research and Ethics Committee (MREC), Ministry of Health Malaysia. Patient consent was waived for this study due to the use of cultured isolates from retrospective samples and data were analysed anonymously. All methods were carried out in accordance with Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mohd Abd Razak, M.R., Md Jelas, N.H., Norahmad, N.A. et al. In vitro study on efficacy of SKF7®, a Malaysian medicinal plant product against SARS-CoV-2. BMC Complement Med Ther 24, 333 (2024). https://doi.org/10.1186/s12906-024-04628-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12906-024-04628-6