Abstract
Background
Upland cotton (Gossypium hirsutum L.) is the most economically important species in the cotton genus (Gossypium spp.). Enhancing the cotton yield is a major goal in cotton breeding programs. Lint percentage (LP) and boll weight (BW) are the two most important components of cotton lint yield. The identification of stable and effective quantitative trait loci (QTLs) will aid the molecular breeding of cotton cultivars with high yield.
Results
Genotyping by target sequencing (GBTS) and genome-wide association study (GWAS) with 3VmrMLM were used to identify LP and BW related QTLs from two recombinant inbred line (RIL) populations derived from high lint yield and fiber quality lines (ZR014121, CCRI60 and EZ60). The average call rate of a single locus was 94.35%, and the average call rate of an individual was 92.10% in GBTS. A total of 100 QTLs were identified; 22 of them were overlapping with the reported QTLs, and 78 were novel QTLs. Of the 100 QTLs, 51 QTLs were for LP, and they explained 0.29–9.96% of the phenotypic variation; 49 QTLs were for BW, and they explained 0.41–6.31% of the phenotypic variation. One QTL (qBW-E-A10-1, qBW-C-A10-1) was identified in both populations. Six key QTLs were identified in multiple-environments; three were for LP, and three were for BW. A total of 108 candidate genes were identified in the regions of the six key QTLs. Several candidate genes were positively related to the developments of LP and BW, such as genes involved in gene transcription, protein synthesis, calcium signaling, carbon metabolism, and biosynthesis of secondary metabolites. Seven major candidate genes were predicted to form a co-expression network. Six significantly highly expressed candidate genes of the six QTLs after anthesis were the key genes regulating LP and BW and affecting cotton yield formation.
Conclusions
A total of 100 stable QTLs for LP and BW in upland cotton were identified in this study; these QTLs could be used in cotton molecular breeding programs. Putative candidate genes of the six key QTLs were identified; this result provided clues for future studies on the mechanisms of LP and BW developments.
Similar content being viewed by others
Background
Cotton (Gossypium) is an economically important natural fiber plant. Upland cotton (Gossypium hirsutum) is the most widely cultivated cotton variety, accounting for approximately 95% of global cotton production [1, 2]. Increasing the yield of upland cotton remains the main objective of this important cash crop worldwide. Cotton yield is typically affected by several complex quantitative traits, including the boll number (BN), lint percentage (LP), boll weight (BW), seed index (SI) and lint index (LI) [3]. These yield component traits are controlled by genetic factors and are affected by environmental factors; they are also genetically related to each other [3,4,5]. LP is an economically important index for cotton cultivars with the highest heritability [6]. Because LP is a key contributor to lint yield and is easy to measure, selection for increasing LP has become an important approach for enhancing lint yield [7, 8]. Numerous studies have shown that cotton yield mainly depends on LP, BW, and BN, and these traits have been positively selected in cultivated cotton throughout the domestication process [9,10,11,12,13,14,15].
Because cotton breeding requires excellent germplasm, a large amount of germplasm resources have been preserved and improved in China, such as many high LP cultivars/lines [16,17,18]. Many interspecific introgressive lines (ILs) or chromosome segment introgression lines (CSILs) have been obtained by crosses between G. hirsutum and Gossypium barbadense [19, 20]; some of these lines have high LP and BW [21]. Many new germplasm resources and cultivars have been successfully bred [22,23,24,25,26]. Our lab has also bred a set of advanced cotton lines/cultivars, such as the parents used in this study.
The identification of stable and effective quantitative trait loci (QTLs) is prerequisites for cotton molecular breeding. From 1998 to 2015, a total of 327 QTLs for LP and 170 QTLs for BW were identified on different chromosomes through meta-QTL analysis [27]. Following the release of the cotton genome sequence, the number of discovered QTLs is rapidly increasing via genome-wide association study (GWAS) or linkage mapping [28,29,30]. For example, structural variations have been explored by resequencing 1,081 G. hirsutum accessions, and 446 structural variations are significantly associated with seven traits, including 21 with LP and 17 with BW [31]. Genetic linkage analysis and association analysis (AS, or GWAS) are the two major approaches for identifying QTLs in crops. Many high-density genetic linkage maps and association maps for cotton have been published. For example, more than 17 crosses or populations of upland cotton have been used to construct genetic maps, including crosses of Yumian1 × T586 [4, 32, 33], Yumian1 × Zhongmiansuo35 [1], NC05AZ06 × NC11-2091 [34], DH962 × Jimian5 [35,36,37], Zhongmiansuo12 (ZMS12) × 8891 [4], (Simian3 × Sumian12) × (Zhong4133 × 8891) [3], Baimian1 × TM-1 [38, 39], Xiangzamian2 [40, 41], HS46 × MARCABUCAG8US-1–88 [42, 43], and CCRI35 × Nan Dan Ba Di Da Hua (NH) [44]. One high-density bin linkage map contains 6,187 bin markers spanning 4,478.98 cM with an average distance of 0.72 cM [18]. Different types of GWAS, including single-locus-GWAS (SL-GWAS), multi-locus GWAS (ML-GWAS), and restricted two-stage, multi-locus, and multi-allele GWAS (RTM-GWAS) approaches, have been used to identify quantitative trait nucleotides (QTNs) for LP and BW in several cotton accessions. More than 16 association maps and many candidate genes for agronomic traits have been reported [5, 8, 10, 12, 45,46,47,48]. For example, 86 single-nucleotide polymorphism linkage disequilibrium block (SNPLDB) loci for LP and 70 SNPLDB loci for BW have been identified from 315 cotton accessions using RTM-GWAS [12]. A total of 719 upland cotton accessions have been screened by GWAS using the cottonSNP63K array, and 62 identified single nucleotide polymorphism (SNP) loci were significantly associated with different traits; a total of 689 candidate genes were screened, and 27 of them contain at least one significant SNP, including three for LP and six for BW [5].
Although the inheritance, QTLs and candidate genes of LP and BW in upland cotton have been widely studied, only a few of the studied QTLs have been used in the molecular breeding of cotton via marker-assisted selection (MAS) [49, 50]. One of the reasons is that the identified QTLs are unstable in multiple-environments and only explain little phenotypic variance. Consequently, mining stable, effective LP and BW-related QTLs or QTNs would greatly aid cotton molecular breeding. We have previously bred the excellent cotton lines ZR014121 and EZ60 and the cultivar CCRI60. Here, we identified stable, effective LP and BW-related QTLs to aid the utilization of the germplasm resources in cotton breeding.
Results
Phenotypic variation in LP and BW
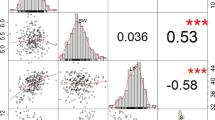
We evaluated two yield-related traits LP and BW, in the two recombinant inbred line (RIL) populations under four environments in 2020 and 2021. The LP and BW ranged from 32.56% to 48.26% and from 4.09 to 6.93 g in P-EZ60, respectively (Table 1); LP and BW ranged from 31.57% to 48.02% and from 3.68 to 6.83 g in P-CCRI60, respectively (Table 2). All of the absolute skewness values of LP and BW were less than 1.0. The distributions of the LP and BW in the four experimental environments were normal. This suggests that LP and BW are polygenic traits, and the data could be used to map QTLs (Fig. 1). LP and BW exhibited high degrees of phenotypic variation. The coefficient of variation for each trait was relatively consistent among the different environments, suggesting that LP and BW were significantly affected by the environment, and the effect on BW (average 7.16 in P-EZ60; 7.55 in P-CCRI60) was greater than that on LP (average 5.69 in P-EZ60; 5.51 in P-CCRI60) (Tables 1, and 2).
The correlations between LP and BW of all the RILs in the four environments were analyzed separately. Generally, LP and BW were significantly negatively correlated in P-EZ60 and P-CCRI60, and the coefficients ranged from -0.098 to -0.340, which suggested that it was difficult to improve LP and BW synchronously (Tables 3, and 4). Because the cotton field was waterlogged in Anyang in 2021, the LP and BW were affected to some extent, but the phenotypic data met the requirements for GWAS (Fig. 1). Analysis of variance (ANOVA) showed that there were highly significant differences among the accessions and environments for the two traits of two populations (Table 5). It indicated that LP and BW were significantly influenced by the accessions and planting environments.
SNP quality control and in silico mapping
According to the high-throughput whole-genome sequencing data of upland cotton (Nanjing Agricultural University), a liquid SNP array with 10 K SNPs was developed. The two RIL populations of P-CCRI60 and P-EZ60, including their parents, were genotyped by genotyping by target sequencing (GBTS) (Table S1). The total number of samples was 500. The average call rate of a single locus was 94.35%, and the average call rate of an individual was 92.10%. The results of the genotype control are shown in supplementary table 2 (Table S2). The BLAST alignment tool was used to analyze the probe sequences of SNPs against the G. hirsutum TM-1 genome sequence [28, 51], and a total of 8,348 genotyped high-quality SNPs across the 500 samples were used in association mapping.
Genome-wide association studies
We used the genetic model of 3VmrMLM to detect QTNs for LP and BW × environment interactions (Fig. 2). A total of 104 stable quantitative trait nucleotides (QTNs) on 26 chromosomes were identified as significantly associated with LP and BW (Table S3). Following other similar studies [47], we defined the flanking 200-Kb regions of QTNs as an initial QTL and merged the overlapping QTLs to obtain the final QTLs. In the end, 100 stable QTLs were detected; 51 of them were for LP and 49 were for BW, including three QEIs, one for LP and two for BW, which could be identified in the four environments (Table S4). A total of 20 stable QTLs, 14 for LP and 6 for BW, were identified in EZ60, including one QEI for BW that could be identified in the four environments; 33 stable QTLs, 18 for LP and 15 for BW, were identified in CCRI60, including one QEI for LP that could be identified in the four environments; and 47 stable QTLs were identified in ZR014121, 19 for LP and 28 for BW, including one QEI for BW that could be identified in the four environments (Table S4). One QTL in chromosome A10, qBW-E-A10-1, was identified in both populations. Among the 100 QTLs, 22 QTLs, 9 for LP and 13 for BW, were overlapping with the reported QTLs (Table S5); 78 QTLs, 42 for LP and 36 for BW, were novel (Table S6).
Manhattan-plots of LP and BW using the genetic model 3VmrMLM. X-axes are cotton chromosomes. Y-axes on the left side report -log10 P-values of the main-effect QTNs, which were obtained from single-marker genome-wide scans for all the markers in the first step of 3VmrMLM; Y-axes on the right side report LOD scores, which were obtained from likelihood ratio tests for significant and suggested QTNs, with a threshold of LOD = 3.0 (dashed line) in the second step of 3VmrMLM. These LOD scores are indicated by points with straight lines
The QTLs explained 0.29–9.96% of the phenotypic variations in LP or BW. In P-EZ60, the novel QTLs associated with LP explained 0.47–8.67% of the phenotypic variation, and the novel QTLs associated with BW explained 0.91–6.31% of the phenotypic variation. In P-CCRI60, the novel QTLs associated with LP explained 0.29 –9.96% of the phenotypic variation, and the novel QTLs associated with BW explained 0.36–3.02% of the phenotypic variation.
In sum, a total of 51 QTLs related to LP were detected in this study, including 14 in EZ60, 18 in CCRI60, and 19 in ZR014121; 28 QTLs were in the At subgenome, and 27 QTLs were in the Dt subgenome, indicating that LP-related QTLs were evenly distributed in the At and Dt subgenomes. A total of 49 QTLs related to BW were detected, including 6 in EZ60, 15 in CCRI60, and 28 in ZR014121; 34 QTLs were in the At subgenome, and 15 QTLs were in the Dt subgenome, indicating that the QTLs related to BW were mainly distributed in the At subgenome. There were two QEIs, which were located on chromosomes A02 and A10 (Fig. 3).
Candidate genes in the regions of the six key QTLs
To identify candidate genes of key QTLs, six QTLs were selected, including three QEIs, the common QTL qBW-E-A10-1 that was mapped in both populations and two important QTLs (qLP-E-D03-2 and qLP-C-D03-2). The three QEIs were QTLs that were stable in the four environments (Table S7). A total of 108 putative candidate genes in the regions of the six key QTLs in multiple environments were identified, including genes that were positively related to LP and BW, such as the genes involved in gene transcription, protein synthesis, calcium signaling, phytohormone synthesis and signaling, and fiber synthesis-related polysaccharide metabolism (Table S6).
KEGG analysis showed that the 48 genes related to LP were mainly involved in “metabolic pathways” and “spliceosome” (Fig. 4). Eighteen metabolic pathways such as “biosynthesis of secondary metabolites”, “microbial metabolism in diverse environments” and “DNA replication” were also detected. KEGG analysis showed that the 60 genes related to BW were mainly involved in “metabolic pathways” and “biosynthesis of secondary metabolites” (Fig. 5). “Microbial metabolism in diverse environments”, “carbon metabolism,” “glycolysis/gluconeogenesis,” and 19 other metabolic pathways were detected.
A histogram of candidate genes enriched in different KEGG pathways for LP. The x-axis indicates the number of candidate genes. The y-axis represents biological processes. The details are listed in Table S9
A histogram of candidate genes enriched in different KEGG pathways for BW. The x-axis indicates the number of candidate genes. The y-axis represents biological processes. The details are listed in Table S10
Expression profiles of candidate genes during fiber development
Most of the candidate genes associated with LP and BW were differentially expressed in cotton fiber at different developmental stages, and there were differences at expression levels between the high-LP parent EZ60 and the low-LP parent ZR014121 at the same stage (Fig. 6). Among the major candidate genes, Gh_A02G0096 was only expressed in the ovule developmental stage of EZ60. Gh_A02G0111 was mainly expressed in both EZ60 and ZR014121 at 0, 5, 10, and 20 days post-anthesis (DPA). Its expression levels were higher in ZR014121 than in EZ60 at 0, 5, and 25 DPA; its expression levels were higher in EZ60 than in ZR014121 at 10 DPA. Gh_D03G1064 was highly expressed in both EZ60 and ZR014121 at all stages. It was mainly expressed at 0, 5, and 10 DPA, and its expression level in ZR014121 was higher than that in EZ60 at 10 DPA. Gh_D03G1069 was expressed in both EZ60 and ZR014121 at all stages. Its expression levels were higher in ZR014121 than in EZ60 at 10 and 20 DPA; its expression levels were higher in EZ60 than in ZR014121 at 0, 5, 15, and 25 DPA. Gh_A02G0106 was significantly highly expressed during the ovule development stage in EZ60, highly expressed at 5 DPA, and weakly expressed at 10 DPA in ZR014121.
Gene expression profiles of the candidate genes of LP and BW QTLs during fiber development in EZ60 and ZR014121. Each column represents one sample, and rows represent candidate genes. The expression levels of the candidate genes (FPKM) were log2-normalized (i.e., log2(FPKM + 0.01)) and presented in different colours on the scale bar. ZR indicates cotton line ZR014121; DPA indicates days post-anthesis. 0 DPA represents the ovule development stage. 5, 10, 15, 20, and 25 DPA represent the fiber development stages. Detailed information on gene expression is shown in Table S11
Co-expression of candidate genes
The interaction network of candidate genes associated with LP and BW was investigated by constructing the protein–protein interaction (PPI) network using the STRING database [52] (Fig. 7). Correlations were observed in the expression of the following proteins that appear to comprise a co-expression network: Gh_A02G0111, Gh_D03G1056, Gh_D03G1134, Gh_D03G1064, Gh_A02G0106, Gh_A10G1521, and Gh_A10G1653. Network analysis of the major proteins was carried out using Cytoscape 3.7.2 (Fig. 8). Gh_D03G1056, Gh_D03G1064, Gh_D03G1134, and Gh_A02G0111 played important roles in the network.
Protein–protein interaction of the candidate genes of the QTLs for LP and BW. Network nodes represent proteins with splice isoforms or post-translational modifications are collapsed, i.e. each node represents all the proteins produced by a single, protein-coding gene locus. Colored nodes: query proteins and first shell of interactors; white nodes: second shell of interactors; Empty nodes: unknown proteins. 3D structure filled nodes: some 3D structures are known or predicted. Edges represent protein–protein associations. Associations are meant to be specific and meaningful (i.e., proteins jointly contribute to a shared function); this does not necessarily mean that they physically bind to each other. Known Interactions, blue: from curated databases; purple: experimentally determined. Predicted Interactions, green: gene neighborhood, red: gene fusions; indigo: gene co-occurrence; Others, yellow: textmining, black: co-expression, light purple: protein homology
Major gene coexpression network of the candidate genes of the QTLs for LP and BW. Lines indicate co-expression of two linked genes. Network nodes represent genes. The size of the circle shows the betweenness centrality points of the gene. The size of the circle indicates that the gene plays an important role in co-expression. In this graph, genes with higher betweenness centrality points are marked in green and placed in the outer circle, and genes with smaller BC values are marked in red and placed in the inner circle. The three genes in the outer ring, Gh_D03G1056, Gh_D03G1064, and Gh_D03G1134 were candidate genes for LP, and Gh_A02G0111 was a candidate gene for BW
PPI analysis indicated that GAI interacted with six other proteins. GAI interacted with FRI; FRI interacted with FPA; FOA interacted with AT1G12775; AT1G12775 interacted with AT3G46960; and AT3G46960, AT3G06700, and AT1G80750 interacted with each other (Fig. 7). There were three groups of co-expressed genes, UBC32 and PCNA1; and CRT3 and ECA1; HCF107 and GOX1. Co-expression analysis of the 108 candidate genes of the six QTLs using Cytoscape 3.7.2 indicated that the seven genes (the same as the result of PPI) were co-expressed, including Gh_A02G0106 (GAI) (Fig. 8).
Discussion
A set of new major QTLs for LP and BW that could be used for MAS was obtained
LP and BW are the most important traits in cotton breeding, and they have been widely studied. More than 417 unique QTLs for LP have been identified on 26 chromosomes, including 243 QTLs identified with LOD > 3. More than 60 were stable, major effective QTLs that could be used for MAS [50]. According to the CottonGen Database [53, 54], a total of 1,387 yield QTLs and four yield component trait QTLs have been identified. The numbers of these QTLs are increasing continually. Recently, 34 SNPs corresponding to 22 QTLs for LP, including 13 novel QTLs, were detected from 254 upland cotton accessions via GWAS [55]. Two stable LP QTLs and three BW QTLs were identified in the RIL mapping population derived from the inter-specific cross between G. hirsutum cv DS-28 and G. barbadense cv SBYF-425 [56]. We also identified one QTL for LP, and nine QTLs for BW from a BC5F3:5 chromosome segment substitution line population derived from G. hirsutum CCRI36 and G. barbadense Hai1 [57]. Three QTLs for LP and one QTL for BW were identified from an F2 population derived from the G. hirsutum × G. barbadense cross [58].
In this study, a total of 51 stable QTLs for LP and 49 stable QTLs for BW were identified from three upland cotton lines ZR014121, CCRI60, and EZ60; these QTLs could explain 0.29–9.96% of the phenotypic variation in LP and 0.41–6.31% of the phenotypic variation in BW. A total of 78 of these QTLs were novel. These findings enhance QTL resources that could be used to enhance the yield of cotton; this QTL information will also aid the molecular breeding of cotton cultivars with high yield.
Many studies have shown that the heritability of LP is the highest among all yield component traits in cotton, and the heritability of BW was the lowest among all cotton yield components. Because the heritability of BW is low, environmental factors can have significant effects on BW [6, 59,60,61]. The results of this study also demonstrate that environmental factors have stronger effects on BW than on LP (Tables 1, and 2). Thus, selection for LP can achieve desired outcomes more efficiently than selection for BW in cotton breeding. Correlations and path analysis among agronomic and technological traits of 16 upland cotton lines indicated that LP was negatively correlated with BW (-0.2668) [62]. Generally, LP and BW are negatively related [50]. In our study, the correlation coefficients between LP and BW ranged from -0.098 to -0.340 (Tables 3, and 4). This indicates that increases in one of these traits limit increases in the other. LP may be the target of direct selection on cotton genotypes with high cotton fiber yield.
Most QTLs for LP and BW explain less than 10% of the phenotypic variation. For example, one study indicates that nine QTLs for LP explain 1.84–13.50% of the observed phenotypic variation; two QTLs for BW explain 6.02–9.50% of the observed phenotypic variation [63]. The QTLs qLP-C13-1 and qLP-C25-1 for LP explain 5.77% and 8.87% of the phenotypic variation, respectively [64]. A GWAS of a set of 289 Gossypium arboreum chromosome segment ILs in G. hirsutum indicates that co-QTLs for LP explain 1.21–10.79% of the phenotypic variation, and co-QTLs for BW explain 1.17–11.56% of the phenotypic variation [65]. Some QTLs for LP identified in this study explained nearly 10% of the phenotypic variation, and all QTLs for BW explained less than 10% of the phenotypic variation (Table S4). These QTLs, especially the major effective QTLs, can be used to breed cotton plants with high yield via MAS.
Several putative candidates of the six QTLs for LP and BW were identified
Understanding the molecular mechanisms of LP and BW developments is essential for the molecular breeding of cotton plants with high yield, especially via genetic engineering. Many candidate genes of the QTLs for LP and BW have been studied [48,49,50, 55]. The TIP41-like family protein (TIP41L) gene (GH_A12G0194) is thought to be the candidate gene of a stable major QTL (q(BW + SI)-A12-1) for BW [49]. One gene orthologous to the Arabidopsis receptor-like protein kinase gene HERK1 (GB_A07G1034) was predicted to be the candidate gene for LP in G. barbadense [48]. Two candidate genes (Gh_D01G0162 and Gh_D07G0463) of QTLs for LP were identified. Gh_D01G0162 is a homolog of the auxin-responsive GH3 family protein gene, and Gh_D07G0463 is a homolog of the NADPH/respiratory burst oxidase protein D gene (RBOHD) in Arabidopsis [55]. A molecular regulatory network for LP has been proposed based on the functions of the candidate genes of QTLs for LP [50].
In this study, the candidate genes of the six important QTLs for LP and BW were investigated. The QTLs for both traits have candidate genes involved in gene transcription, protein syntheses, signaling, calcium signaling, carbon metabolism, metabolic pathways, and biosynthesis of secondary metabolites, which demonstrates that there are several candidate genes of the QTLs for LP and BW (Figs. 4, and 5; Tables S8, S9, S10). This result is consistent with the findings of previous studies [48, 50, 55, 66, 67]. The difference is that a greater number of candidate genes in QTLs for LP were involved in gene expression processes, and a greater number of candidate genes in QTLs for BW were involved in metabolic pathways. Interaction network analysis of the candidate genes associated with LP and BW indicated that seven candidate genes could form a co-expression network. The candidate gene Gh_A02G0096 of qBW-E-A02-1 encodes a homolog of eukaryotic translation initiation factor 2A, and the candidate gene Gh_D03G1069 of qLP-E-D03-2 likely encodes a serine/threonine-protein kinase. Their interaction suggests that LP and BW are closely related during development (Figs. 7, and 8). Additional studies are needed to clarify why LP and BW are negatively related.
Many candidates of the six QTLs are involved in fiber development
The MYB-bHLH-WD40 (including MYB-DEL-TTG and CPC-MYC-TTG) [33, 68] and TCP-HOX-HD [66, 69] regulatory complexes play key roles in cotton fiber development. Phytohormone balance, Ca2+ signaling, and ROS also play key roles regulating fiber development [50, 70, 71].
Many candidate genes of the QTLs for LP and BW are involved in various signaling pathways and metabolic processes in this study, such as the transcription factor bHLH113 gene (Gh_A02G0095); Ca2+ signaling genes (Gh_A10G1519, Gh_D03G1058, and Gh_D03G1266); protein kinase genes (Gh_D03G1144, Gh_D03G1264, and Gh_D03G1069); GA signaling genes (Gh_A02G0104 and Gh_A02G0106); and ROS metabolism-related genes (Gh_D03G1138, Gh_D03G1063, and Gh_D03G1062) [55] (Table S7). Gh_D03G1264 encodes a HERK1-like protein [48]. Gh_A02G0106 is a homolog of AT1G14920, that encodes a gibberellin insensitive protein (DELLA protein GAI), and plays a role in seed germination [72]. Gh_A02G0111 is a homolog of AT2G43410, which encodes a flowering time control protein FPA in Arabidopsis [73]. Gh_D03G1064 encodes a FRIGIDA-like protein that can pleiotropically increase lint yield; it is also significantly associated with SI [5]. The homologous gene of Gh_D03G1064 in Arabidopsis is FRI (AT4G00650), which regulates flowering time in Arabidopsis [73,74,75,76,77].
GhFSN1 is a cotton NAC transcription factor that acts as a positive regulator to control secondary cell wall (SCW) formation in cotton fibers by activating downstream SCW-related genes, including GhDUF231L1, GhKNL1, GhMYBL1, GhGUT1 and GhIRX12 [66]. The candidate gene Gh_A02G0101 also encodes a NAC protein (Table S7). The glucosyltransferases, Rab-like GTPase activators, and myotubularin (GRAM) domain gene GhGRAM31 (Ghir_D02G018120) regulate fiber elongation. GhGRAM31 directly interacts with GhGRAM5 and GhGRAM35. GhGRAM5 also interacts with the transcription factor GhTTG1, and GhGRAM35 interacts with the transcription factors GhHOX1 and GhHD1 [67]. The candidate gene Gh_A02G0094 also encodes the C2 and GRAM domain-containing protein At1g03370 (Table S7).
The above data demonstrate that most of the putative candidates of the six QTLs for LP and BW identified in this study were involved in regulating cotton fiber development. Most of the data obtained in this research are consistent with the findings of other studies, indicating that our results were reliable.
Candidate gene expression profiles determine LP and BW
ZR014121 is an excellent high-yield but low-LP line. EZ60 is an early maturity line with high LP. The candidate gene expression profiles of the six QTLs for LP and BW in the two lines significantly differed (Fig. 6). Most candidate genes were highly expressed at the ovule developmental stage (0 DPA) in both ZR014121 and EZ60. Four key candidate genes were highly expressed at 5 DPA in ZR014121, including Gh_A02G0095 (BHLH113, which might be involved in MYB-bHLH-WD40 complexes [33, 68]), Gh_A02G0097 (RGA3), Gh_A10G1158 (CBDAS), and Gh_D03G1062 (RBOHC, which might be involved in ROS [70]). Gh_A02G0114 (ccdc94) was significantly highly expressed at 15 DPA in EZ60. Gh_A02G0101 (NAC014, which might be involved in SCW formation in cotton fibers [66]) was significantly highly expressed at 25 DPA in ZR014121.
Most genes were highly expressed at the ovule developmental stage, which demonstrates that these genes were highly active in this stage. The expression of four genes in ZR014121 after this stage was likely the main cause of high yield. These four genes, in addition to the other two highly expressed genes, Gh_A02G0114 and Gh_A02G0101, were the key candidate genes of the six QTLs for LP and BW (Fig. 6). Although we were unable to determine whether the six genes represent the six QTLs, our findings indicate that they are the key genes regulating LP and BW and thus affecting cotton yield. These genes provide important genetic resources for studies of the lint regulation mechanism and improvements in cotton yield.
Conclusions
Two RIL populations were constructed using the three excellent upland cotton lines ZR014121, CCRI60, and EZ60, which differ in fiber yield and quality traits. The RILs were genotyped by GBTS and phenotyped under four different environments; a GWAS was then conducted to identify useful yield-related QTLs. A total of 51 QTLs for LP and 49 QTLs for BW were identified, and these QTLs could explain 0.29–9.96% of the phenotypic variation in LP and 0.41–6.31% of the phenotypic variation in BW. There were six major and effective QTLs, three for LP and three for BW, and these could be used to breed cotton with high yield via molecular breeding approaches. A total of 108 putative candidate genes were identified in the six key QTLs, including genes that were positively related to the development of LP and BW, such as genes involved in gene transcription, protein synthesis, calcium signaling, phytohormone synthesis and signaling, and fiber synthesis-related polysaccharide metabolism. Seven of the candidate genes form a co-expression network. Six significantly highly expressed candidate genes after anthesis were important factors regulating cotton yield. These candidate genes will help clarify the molecular mechanisms underlying variation in LP and BW.
Methods
Plant materials and growth conditions
Three G. hirsutum lines ZR014121, CCRI60, and EZ60 were used as parents in this study, and they were bred at the Institute of Cotton Research, Chinese Academy of Agricultural Sciences. All of the three RIL lines we were authorized to use. EZ60 and ZR014121 were preserved in the National Germplasm Library (38 Huanghe Avenue, Anyang, Henan 455,000); their accession numbers were M116025 and ZM115357, respectively. CCRI60 is a variety. ZR014121 has high yield but low LP. EZ60 is an early maturity line with high LP. CCRI60 is an excellent cultivar with several desirable agronomic traits. Two RIL populations at the F6:8 generation in 2020 (at F6:9 in 2021), P-CCRI60 and P-EZ60 were constructed from crosses of ZR014121 × CCRI60 and ZR014121 × EZ60, respectively. P-CCRI60 consisted of 300 RILs, and P-EZ60 consisted of 200 RILs.
There were four factors in the field experiment: two years (2020 and 2021) and two locations (Anyang (36°05′N, 114°29′E), Henan Province, and Weixian (37°58′N, 115°16′E), Hebei Province, China(both of them are our experimental field)); these were each referred to as 20AY, 20WX, 21AY, and 21WX. To eliminate field effects, the experiment was conducted in a randomized incomplete block design with two replicates of each environmental factor. The parents and RILs were planted in rows with lengths of 3 m and widths of 0.8 m; the one control, CCRI60, had 20 rows. The lines were planted in April and sampled in September each year. Field management techniques followed those of regular breeding practices.
Trait measurements
Two yield-related traits LP and BW were evaluated at each field location. The samples were prepared around September 20 each year. Thirty naturally opened bolls from the central part of plants (two bolls on each plant) of each line were randomly hand-harvested to calculate the BW (g) and gin the fiber. Fiber samples were separately weighed to calculate the LP (%). All statistical analyses, including correlations between traits, analysis of variance and significance analyses were conducted using IBM SPSS 22.0 software.
GBTS
For genotyping, the young leaf tissues of the three parents ZR014121, CCRI60, and EZ60, and the RILs of the two populations, P-CCRI60 and P-EZ60, were sampled in July 2020. Genomic DNA was extracted from each sample using a modified cetyltrimethylammonium bromide method [78].
For GBTS, we used the Allegro Targeted Genotyping of NuGEN Technologies; the stable markers covering whole cotton genomes were selected from known markers obtained from the high-throughput sequencing results. To prevent the 3′-ends of the probes from overlapping with other known variable sites, the SNPs were tested in the parents and their F1 plants, and the polymorphic SNPs were used to design primers. DNA fragmentation, adapter ligation, target extension, and library amplification were performed following the instructions of various kits (NuGEN Technologies, San Carlos, California, USA). The libraries were tested using the most recently updated Illumina manufacturer’s instructions (Illumina, San Diego, CA, USA). Three replications of GBTS were performed on each sample.
After the SNP data were generated by BCFtools, the raw SNPs and Indels were screened using three parameters QUAL, RPB, and AC [(-e ‘%QUAL < 100); (RPB < 0.1, %QUAL < 100); (AC < 2, %QUAL < 100)’)]. The cover rate of each sequenced SNP was statistically analyzed using ‘samtools depth’. The SNPs with sequencing cover rates more than 10 times and without genotypes were considered to be genotypes consistent with those in the cotton reference genome; SNPs with sequencing cover rates less than one time and without genotypes were referred to as deletion genotypes. The two SNP quality control criteria were (1) call rate of a single locus and (2) call rate of an individual. The Perl soft program that we translated and edited was used to statistically analyze the quality control criteria. For the physical localizations of the SNP markers, the probe sequences of the SNPs were used to| perform local BLAST [79] queries against the G. hirsutum TM-1 reference genome [28, 52].
GWAS
The high-quality SNPs determined from the whole study populations, P-CCRI60 and P-EZ60, were used to conduct a GWAS for LP and BW. Given the possibility of obtaining false-positive QTNs with low association frequencies, we selected QTNs with LOD > 3 as stable QTNs in subsequent analyses. The software 3VmrMLM version 1.0 [80] was used to perform GWAS with the following settings: method = ‘Multi_env’; fileKin = NULL; filePS = NULL; PopStrType = ‘Q’; fileCov = NULL; SearchRadius = 20; svpal = 0.01; DrawPlot = TRUE; Plotformat = ‘pdf’; and Chr_name_com = NULL. We obtained significant and suggested main-effect QTNs, significant, as well as suggested QEIs. The significant QTNs were selected by Bonferroni correction, and the critical P-value was 0.05/m, where m is the number of tests or markers, and suggested QTNs were identified as those with LOD ≥ 3.0. Significant QEIs were selected by Bonferroni correction; the critical P-value was 0.05/m, where m is the number of tests or markers, and suggested QEIs were identified as those with LOD ≥ 3.0 using default parameters [80].
Prediction and identification of candidate genes
We defined the flanking 200-Kb regions of the QTNs as the same QTL and merged the overlapping QTLs to confirm the number of QTLs [81]. Potential candidate genes were confirmed based on gene annotations in the G. hirsutum TM-1 genome [28, 52]. All the candidate genes were subjected to Gene Ontology [82] enrichment analysis and Kyoto Encyclopedia of Genes and Genomes [83,84,85] analysis. The interaction network of candidate genes was inferred by constructing a PPI network using the STRING database [52]. The network analysis was conducted using Cytoscape 3.7.2.
RNA sequencing and gene expression profiles of the QTL candidates
The ovules/fibers of EZ60 and ZR014121 were sampled at 0, 5, 10, 15, 20, and 25 days post-anthesis (DPA). The total RNAs were extracted using the mirVana™ miRNA Isolation Kit (Ambion) according to the manufacturer’s instructions. Three biological replicates were performed for each sample. The Illumina PE libraries were sequenced on the HiSeqTM2500 (Illumina) platform. Raw reads were filtered using Trimmomatic-0.39 [86], and the clean reads were mapped to the reference genome [87] using STAR-2.7.9a [88]; the abundances of transcripts were quantified using RSEM-1.2.26 [89]. Differentially expressed genes (DEGs) were identified using DESeq2-1.30.1 according to the following criteria: padj < 0.05 and log2 (FoldChange) > 1 DESeq2-1.30.1 [90]. Hierarchical cluster analysis of DEGs was conducted to measure expression levels. The expression profiles of every candidate gene were used to preliminarily identify LP-related and BW-related genes.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the NCBI repository, [ https://www.ncbi.nlm.nih.gov/bioproject/906276] [Accession number: PRJNA906276].
References
Chen ZJ, Scheffler BE, Dennis E, Triplett BA, Zhang T, Guo W, Chen X, Stelly DM, Rabinowicz PD, Town CD, Arioli T, Brubaker C, Cantrell RG, Lacape JM, Ulloa M, Chee P, Gingle AR, Haigler CH, Percy R, Saha S, Wilkins T, Wright RJ, Van Deynze A, Zhu Y, Yu S, Abdurakhmonov I, Katageri I, Kumar PA, Rahman, M-U, Zafar Y, Yu JZ, Kohel R J, Wendel JF, Paterson AH. Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 2007;145:1303–10.
Imran M, Shakeel A, Azhar FM, Farooq J, Saleem MF, Saeed A, Nazeer W, Riaz M, Naeem M, Javaid A. Combining ability analysis for within-boll yield components in upland cotton (Gossypium hirsutum L.). Genet Mol Res. 2012;11(3):2790–800.
Qin H, Guo W, Zhang YM, Zhang T. QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor Appl Genet. 2008;117:883–94.
Wang B, Guo W, Zhu X, Wu Y, Huang N, Zhang T. QTL Mapping of yield and yield components for elite hybrid derived-RILs in upland cotton. J Genet Genomics. 2007;34(1):35–45.
Sun Z, Wang X, Liu Z, Gu Q, Zhang Y, Li Z, Ke H, Yang J, Wu Ji, Wu L, Zhang G, Zhang C, Ma Z. A genome wide association study uncovers novel genomic regions and candidate genes of yield related traits in upland cotton. Theor Appl Genet. 2018;131(11):2413–25.
Badigannavar A, Myers GO. Genetic diversity, population structure and marker trait associations for seed quality traits in cotton (Gossypium hirsutum). J Genet. 2015;94(1):87–94.
Tang F, Xiao W. Genetic effects and heterosis of within-boll yield components in upland cotton (Gossypium hirsutum L.). Euphytica. 2013;194:45–51.
Su J, Fan S, Li L, Wei H, Wang C, Wang H, Song M, Zhang C, Gu L, Zhao S, Mao G, Wang C, Pang C, Yu S. Detection of favorable QTL alleles and candidate genes for lint percentage by GWAS in Chinese Upland cotton. Front Plant Sci. 2016;7:1576.
Jiang Y, Guo W, Zhu H, Ruan YL, Zhang T. Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol J. 2012;10:301–12.
Fang L, Wang Q, Hu Y, Jia Y, Chen J, Liu B, Zhang Z, Guan X, Chen S, Zhou B, Mei G, Sun J, Pan Z, He S, Xiao S, Shi W, Gong W, Liu J, Ma J, Cai C, Zhu X, Guo W, Du X, Zhang T. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat Genet. 2017;49(7):1089–98.
Song C, Li W, Pei X, Liu Y, Ren Z, He K, Zhang F, Sun K, Zhou X, Ma X, Yang D. Dissection of the genetic variation and candidate genes of lint percentage by a genome-wide association study in upland cotton. Theor Appl Genet. 2019;132(7):1991–2002.
Su J, Wang C, Ma Q, Zhang A, Shi C, Liu J, Zhang X, Yang D, Ma X. An RTM-GWAS procedure reveals the QTL alleles and candidate genes for three yield related traits in upland cotton. BMC Plant Biol. 2020;20(1):416.
Zhang Z, Li J, Jamshed M, Shi Y, Liu A, Gong J, Wang S, Zhang J, Sun F, Jia F, Ge Q, Fan L, Zhang Z, Pan J, Fan S, Wang Y, Lu Q, Liu R, Deng X, Zou X, Jiang X, Liu P, Li P, Iqbal MS, Zhang C, Zou J, Chen H, Tian Q, Jia X, Wang B, Ai N, Feng G, Wang Y, Hong M, Li S, Lian W, Wu B, Hua J, Zhang C, Huang J, Xu A, Shang H, Gong W, Yuan Y. Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population. Plant Biotechnol J. 2020;18(1):239–53.
Wang F, Zhang J, Chen Y, Zhang C, Gong J, Song Z, Zhou J, Wang J, Zhao C, Jiao M, Liu A, Du Z, Yuan Y, Fan S, Zhang J. Identification of candidate genes for key fibre-related QTLs and derivation of favourable alleles in Gossypium hirsutum recombinant inbred lines with G. barbadense introgressions. Plant Biotechnol J. 2020;18:707–20.
Zhu G, Hou S, Song X, Wang X, Wang W, Chen Q, Guo W. Genome-wide association analysis reveals quantitative trait loci and candidate genes involved in yield components under multiple field environments in cotton (Gossypium hirsutum). BMC Plant Biol. 2021;21:250.
Zeng L, Wu J. Germplasm for genetic improvement of lint yield in Upland cotton: genetic analysis of lint yield with yield components. Euphytica. 2012;187:247–61.
Du XM, Pan JJ, Wang RH, Zhang TZ, Shi YZ. Genetic analysis of presence and absence of lint and fuzz in cotton. Plant Breeding. 2001;120:519–22.
Han Z, Hu Y, Tian Q, Cao Y, Si A, Si Z, Zang Y, Xu C, Shen W, Dai F, Liu X, Fang L, Chen H, Zhang T. Genomic signatures and candidate genes of lint yield and fibre quality improvement in Upland cotton in Xinjiang. Plant Biotechnol J. 2020;18:2002–14.
Shi Y, Li W, Li A, Ge R, Zhang B, Li J, Liu G, Li J, Liu A, Shang H, Gong J, Gong W, Yang Z, Tang F, Liu Z, Zhu W, Jiang J, Yu X, Wang T, Wang W, Chen T, Wang K, Zhang Z, Yuan Y. Constructing a high-density linkage map for Gossypium hirsutum × Gossypium barbadense and identifying QTLs for lint percentage. J Integr Plant Biol. 2015;57(5):450–67.
Li PT, Rashid MHO, Chen TT, Lu QW, Ge Q, Gong WK, Liu AY, Gong JW, Shang HH, Deng XY, Li JW, Li SQ, Xiao XH, Liu RX, Zhang Q, Duan L, Zou XY, Zhang Z, Jiang X, Zhang Y, Peng RH, Shi YZ, Yuan YL. Transcriptomic and biochemical analysis of upland cotton (Gossypium hirsutum) and a chromosome segment substitution line from G. hirsutum × G. barbadense in response to Verticillium dahliae infection. BMC Plant Biol. 2019;19(1):19.
Zhu XF, Wang P, Si ZF, Zhang TZ. QTL mapping for yield components in Gossypium barbadense chromosome segment introgression lines based on Gossypium hirsutum background. Acta Agr Sin (in Chinese). 2017;43(12):1784–90.
Wang SP, Xiao SD. Breeding advance of the new cotton line C24 with high lint percentage. Chin Agric Sci Bull (in Chinese). 1996;12(6):27–8.
Ma X, Wang Z, Li W, Zhang Y, Zhou X, Liu Y, Ren Z, Pei X, Zhou K, Zhang W, He K, Zhang F, Liu J, Ma W, Xiao G, Yang D. Resequencing core accessions of a pedigree identifies derivation of genomic segments and key agronomic trait loci during cotton improvement. Plant Biotechnol J. 2019;17(4):762–75.
Li M, Wang ZZ. Characterization, screening and enhancement of cotton germplasm. Zuo Wu Pin Zhong Zi Yuan (in Chinese). 1992;3:11–2.
Chen Y, Liu G, Ma H, Song Z, Zhang C, Zhang J, Zhang J, Wang F, Zhang J. Identification of introgressed alleles conferring high fiber quality derived from Gossypium barbadense L. in secondary mapping populations of G. hirsutum L. Front Plant Sci. 2018;9:1023.
Zhang Z, Shang H, Shi Y, Huang L, Li J, Ge Q, Gong J, Liu A, Chen T, Wang D, Wang Y, Palanga KK, Muhammad J, Li W, Lu Q, Deng X, Tan Y, Song W, Cai J, Li P, Rashid H, Gong W, Yuan Y. Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum.). BMC Plant Biol. 2005;16:79.
Said JI, Song M, Wang H, Lin Z, Zhang X, Fang DD, Zhang J. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol Genet Genomics. 2015;290(3):1003–25.
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Yu, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–7.
Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, Ye Z, Huang H, Yan F, Ma Y, Zhang L, Liu M, You J, Yang Y, Liu Z, Huang F, Li B, Qiu P, Zhang Q, Zhu L, Jin S, Yang X, Min L, Li G, Chen LL, Zheng H, Lindsey K, Lin Z, Udall JA, Zhang X. Reference genome sequences of two cultivated allotetraploid cottons. Gossypium hirsutum and Gossypium barbadense, Nat Genet. 2019;51:224–9.
Huang G, Wu Z, Percy RG, Bai M, Li Y, Frelichowski JE, Hu J, Wang K, Yu JZ, Zhu Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat Genet. 2020;52:516–24.
Ma Z, Zhang Y, Wu L, Zhang G, Sun Z, Li Z, Jiang Y, Ke H, Chen B, Liu Z, Gu Q, Wang Z, Wang G, Yang J, Wu J, Yan Y, Meng C, Li L, Li X, Mo S, Wu N, Ma L, Chen L, Zhang M, Si A, Yang Z, Wang N, Wu L, Zhang D, Cui Y, Cui J, Lv X, Li Y, Shi R, Duan Y, Tian S, Wang X. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement. Nat Genet. 2021;53(9):1385–91.
Zhang ZS, Xiao YH, Luo M, Li XB, Luo XY, Hou L, Li DM, Pei Y. Construction of a genetic linkage map and QTL analysis of fiber-related traits in upland cotton (Gossypium hirsutum L.). Euphytica. 2005;144:91–9.
Liu D, Liu F, Shan X, Zhang J, Tang S, Fang X, Liu X, Wang W, Tan Z, Teng Z, Zhang Z, Liu D. Construction of a high-density genetic map and lint percentage and cottonseed nutrient trait QTL identification in upland cotton (Gossypium hirsutum L.). Mol Genet Genomics. 2015;290:1683–700.
Zhang K, Kuraparthy V, Fang H, Zhu L, Sood S, Jones DC. High-density linkage map construction and QTL analyses for fiber quality, yield and morphological traits using CottonSNP63K array in upland cotton (Gossypium hirsutum L.). BMC Genomics. 2019;20:889.
Lin ZX, Feng CH, Guo XP, Zhang XL. Genetic analysis of major QTLs and epistasis interaction for yield and fiber quality in upland cotton. Sci Agric Sin (in Chinese). 2009;42(9):3036–47.
Lin ZX, Zhang YX, Zhang XL, Guo XP. A high-density integrative linkage map for Gossypium hirsutum. Euphytica. 2009;166:35–45.
Wang H, Huang C, Zhao W, Dai B, Shen C, Zhang B, Li D, Lin Z. Identification of QTL for fiber quality and yield traits using two immortalized backcross populations in upland cotton. PLoS ONE. 2016;11(12): e0166970.
Xia Z, Zhang X, Liu YY, Jia ZF, Zhao HH, Li CQ, Wang QL. Major gene identification and quantitative trait locus mapping for yield-related traits in upland cotton (Gossypium hirsutum L.). J Integr Agr. 2014;13(2):299–309.
Wang M, Li C, Wang Q. Quantitative trait loci mapping and genetic dissection for lint percentage in upland cotton (Gossypium hirsutum). J Genet. 2014;93:371–8.
Liu R, Wang B, Guo W, Wang L, Zhang T. Differential gene expression and associated QTL mapping for cotton yield based on a cDNA-AFLP transcriptome map in an immortalized F2. Theor Appl Genet. 2011;123:439–54.
Liu R, Wang B, Guo W, Qin Y, Wang L, Zhang Y, Zhang T. Quantitative trait loci mapping for yield and its components by using two immortalized populations of a heterotic hybrid in Gossypium hirsutum L. Mol Breeding. 2012;29:297–311.
Wu J, Gutierrez OA, Jenkins JN, McCarty JC, Zhu J. Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of upland cotton. Euphytica. 2009;165:231–45.
Li C, Dong Y, Zhao T, Li L, Li C, Yu E, Mei L, Daud MK, He Q, Chen J, Zhu S. Genome-wide SNP linkage mapping and QTL analysis for fiber quality and yield traits in the upland cotton recombinant inbred lines population. Front Plant Sci. 2016;7:1356.
Diouf L, Magwanga RO, Gong W, He S, Pan Z, Jia YH, Kirungu JN, Du X. QTL mapping of fiber quality and yield-related traits in an intra-specific upland cotton using genotype by sequencing (GBS). Int J Mol Sci. 2018;19:441.
Huang C, Nie X, Shen C, You C, Li W, Zhao W, Zhang X, Lin Z. Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol J. 2017;15(11):1374–86.
Shen C, Wang N, Huang C, Wang M, Zhang X, Lin Z. Population genomics reveals a fine-scale recombination landscape for genetic improvement of cotton. Plant J. 2019;99(3):494–505.
Zhu G, Gao W, Song X, Sun F, Hou S, Liu N, Huang Y, Zhang D, Ni Z, Chen Q, Guo W. Genome-wide association reveals genetic variation of lint yield components under salty field conditions in cotton (Gossypium hirsutum L.). BMC Plant Biol. 2020;20(1):23.
Yu J, Hui Y, Chen J, Yu H, Gao X, Zhang Z, Li Q, Zhu S, Zhao T. Whole-genome resequencing of 240 Gossypium barbadense accessions reveals genetic variation and genes associated with fiber strength and lint percentage. Theor Appl Genet. 2021;134:3249–61.
Feng L, Su Q, Yue H, Wang L, Gao J, Xing L, Xu M, Zhou C, Yang Y, Zhou B. TIP41L, a putative candidate gene conferring both seed size and boll weight, was fine-mapped in an introgression line of Gossypium hirsutum-Gossypium arboretum. Plant Sci. 2022;317: 111197.
Niu H, Ge Q, Shang H, Yuan Y. Heredity. QTL mapping and candidate gene isolation of cotton lint percentage, Front Genet. 2022;13: 855574.
Hu Y, Chen JD, Fang L, Zhang ZY, Ma W, Niu YC, Ju LZ, Deng JQ, Zhao T, Lian JM, Baruch K, Fang D, Liu X, Ruan YL, Rahman MU, Han JL, Wang K, Wang Q, Wu HT, Mei GF, Zang YH, Han ZG, Xu CY, Shen WJ, Yang DF, Si ZF, Dai F, Zou LF, Huang F, Bai YL, Zhang YG, Brodt A, Ben-Hamo H, Zhu XF, Zhou BL, Guan XY, Zhu SJ, Chen XY, Zhang TZ. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–48.
STRING database: https://cn.string-db.org. Accessed 21 Sept 2022.
CottonGen Database: https://www.cottongen.org. Accessed 21 Sept 2022.
Yu J, Jung S, Cheng CH, Lee T, Zheng P, Buble K, Crabb J, Humann J, Hough H, Jones D, Campbell JT, Udall J, Main D. CottonGen: The community database for cotton genomics, genetics, and breeding research. Plants. 2021;10:2805.
Chen Y, Gao Y, Chen P, Zhou J, Zhang C, Song Z, Huo X, Du Z, Gong J, Zhao C, Wang S, Zhang J, Wang F, Zhang J. Genome-wide association study reveals novel quantitative trait loci and candidate genes of lint percentage in upland cotton based on the CottonSNP80K array. Theor Appl Genet. 2022;135(7):2279–95.
Gowda SA, Katageri IS, Patil RS, Kumar PS, Tiwari GJ, Jena SN, Sawant SV. 63 K and 50 K SNP array based high-density genetic mapping and QTL analysis for productivity and fiber quality traits in cotton. Euphytica. 2022;218:93.
Lu Q, Li P, Yang R, Xiao X, Li Z, Wu Q, Gong J, Ge Q, Liu A, Du S, Wang J, Shi Y, Yuan Y. QTL mapping and candidate gene prediction for fiber yield and quality traits in a high-generation cotton chromosome substitution line with Gossypium barbadense segments. Mol Genet Genomics. 2022;297:287–301.
Si Z, Jin S, Chen J, Wang S, Fang L, Zhu X, Zhang T, Hu Y. Construction of a high-density genetic map and identification of QTLs related to agronomic and physiological traits in an interspecific (Gossypium hirsutum × Gossypium barbadense) F2 population. BMC Genomics. 2022;23:307.
Zeng L, Meredith WR Jr, Gutiérrez OA, Boykin DL. Identification of associations between SSR markers and fiber traits in an exotic germplasm derived from multiple crosses among Gossypium tetraploid species. Theor Appl Genet. 2009;119:93–103.
Santos IG, Teodoro PE, Farias FC, Farias FJC, Carvalho LP, Rodrigues JIS, Cruz CD. Genetic diversity among cotton cultivars in two environments in the State of Mato Grosso. Genet Mol Res. 2017;16(2):16029628.
Rehman A, Mustafa N, Du X, Azhar MT. Heritability and correlation analysis of morphological and yield traits in genetically modified cotton. J Cotton Res. 2020;3:23.
Farias FJC, Carvalho LP, Silva Filho JL, Teodoro PE. Correlations and path analysis among agronomic and technological traits of upland cotton. Genet Mol Res. 2016;15(3):15038239.
Zhai H, Gong W, Tan Y, Liu A, Song W, Li J, Deng Z, Kong L, Gong J, Shang H, Chen T, Ge Q, Shi Y, Yuan Y. Identification of chromosome segment substitution lines of Gossypium barbadense introgressed in G. hirsutum and quantitative trait locus mapping for fiber quality and yield traits. PLoS ONE. 2016; 11 (9), e0159101.
Chen Q, Zhou SJ, Sun KT, Liu JJ, Yuan BT, Wang YP, Wang W, Wang YW, Wang BH, Zhuang ZM. QTL mapping of lint percentage in Gossypium mustelinum introgression lines. Southwest China J Agric Sci (in Chinese). 2019;32(8):1735–9.
Feng L, Chen Y, Xu M, Yang Y, Yue H, Su Q, Zhou C, Feng G, Ai N, Wang N, Zhou B. Genome-wide introgression and quantitative trait locus mapping reveals the potential of Asian cotton (Gossypium arboreum) in improving upland cotton (Gossypium hirsutum). Front Plant Sci. 2021;12: 719371.
Zhang J, Huang GQ, Zou D, Yan JQ, Li Y, Hu S, Li XB. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018;217:625–40.
Ye Z, Qiao L, Luo X, Chen X, Zhang X, Tu L. Genome-wide identification of cotton GRAM family proteins reveals that GRAM31 regulates fiber length. J Exp Bot. 2021;72(7):2477–90.
Shangguan X, Yang Q, Wu X, Cao J. Function analysis of a cotton R2R3 MYB transcription factor GhMYB3 in regulating plant trichome development. Plant Biol. 2021;23:1118–27.
Cao JF, Zhao B, Huang CC, Chen ZW, Zhao T, Liu HR, Hu GJ, Shangguan XX, Shan CM, Wang LJ, Zhang TZ, Wendel JF, Guan XY, Chen XY. The miR319-targeted GhTCP4 promotes the transition from cell elongation to wall thickening in cotton fiber. Mol Plant. 2020;13:1063–77.
Tang WX, Tu LL, Yang XY, Tan JF, Deng FL, Hao J, Guo K, Lindsey K, Zhang XL. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 2014;202(2):509–20.
Cheng Y, Lu L, Yang Z, Wu Z, Qin W, Yu D, Ren Z, Li Y, Wang L, Li F, Yang Z. GhCaM7-like, a calcium sensor gene, influences cotton fiber elongation and biomass production. Plant Physiol Biochem. 2016;109:128–36.
Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-wide analysis of genes targeted by phytochrome interacting factor 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21(2):403–19.
Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I. Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell. 2009;21(10):3185–97.
Emami H, Kempken F. PRECOCIOUS1 (POCO1), a mitochondrial pentatricopeptide repeat protein affects flowering time in Arabidopsis thaliana. Plant J. 2019;100(2):265–78.
Schmalenbach I, Zhang L, Ryngajllo M, Jiménez-Gómez JM. Functional analysis of the Landsberg erecta allele of FRIGIDA. BMC Plant Biol. 2014;14:218.
Liu DX, Rajaby R, Wei LL, Zhang L, Yang ZQ, Yang QY, Sung WK. Calling large indels in 1047 Arabidopsis with IndelEnsembler. Nucleic Acids Res. 2021;49(19):10879–94.
Zhang L, Jimenez-Gomez JM. Functional analysis of FRIGIDA using naturally occurring variation in Arabidopsis thaliana. Plant J. 2020;103(1):154–65.
Allen GC, Flores-Vergara MA, Krasnyanski S, Kumar S, Thompson WF. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 2006;1(5):2320–5.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Li M, Zhang YW, Zhang ZC, Xiang Y, Liu MH, Zhou YH, Zuo JF, Zhang HQ, Chen Y, Zhang YM. A compressed variance component mixed model for detecting QTNs, and QTN-by-environment and QTN-by-QTN interactions in genome-wide association studies. Mol Plant. 2022;15:630–50.
Su Y, Guo A, Huang Y, Wang Y, Hua J. GhCIPK6a increases salt tolerance in transgenic upland cotton by involving in ROS scavenging and MAPK signaling pathways. BMC Plant Biol. 2020;20(1):421.
Gene Ontology (GO; http://www.geneontology.org/GO. Accessed 21 Sept 2022.
Kyoto Encyclopedia of Genes and Genomes: (KEGG; http://www.genome.jp/kegg/KEGG. Accessed 21 Sept 2022.
Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30 ([PMID:10592173]).
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–51 ([PMID:33125081]).
Bolger AM, Lohse M, Usadel B. Trimmomatic: a fexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
The cotton reference genome: https://cottonfgd.org/about/download/assembly/genome.Ghir.ZJU.fa.gz. Accessed 21 Sept 2019.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Dewey CN, Li B. RSEM: accurate transcript quantifcation from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Acknowledgements
We thank the reviewers for comments and suggestions on improving the manuscript.
Funding
This work was supported by funding from the National Key Research and Development Program (2021YFF1000100) and Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Contributions
H.N. performed the experiments, analyzed the data, and drafted the manuscript. M.K and L.H. helped with analysis of the data. H.S., Y.Y and Q.G designed the whole study, revised the manuscript and gave the final approval to the version of the manuscript that is being sent for consideration for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
We complied with all relevant institutional, national and international guide-lines with permissions from State Key Laboratory of Cotton Biology, Key Laboratory of Biological and Genetic Breeding of Cotton, The Ministry of Agriculture, Institute of Cotton Research, Chinese Academy of Agricultural Sciences.
Consent for publication
Not applicable.
Competing interests
The authors declare there are no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. The result of GBTS
Additional file 2:
Table S2. The results of sample genotyping
Additional file 3:
Table S3. The result of 3VmrMLM: QEI
Additional file 4:
Table S4. The identified QTLs
Additional file 5:
Table S5. The identified QTLs overlapped with the reported QTLs
Additional file 6:
Table S6. The identified new QTLs
Additional file 7:
Table S7. All candidate genes of the 6 key QTLs
Additional file 8:
Table S8. Annotations of the candidate genes of the six QTLs for BW and LP
Additional file 9:
Table S9. KEGG annotations of the candidate genes of the QTLs for LP
Additional file 10:
Table S10. KEGG annotations of the candidate genes of the QTLs for BW
Additional file 11:
Table S11. The expression levels of the candidate genes
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Niu, H., Kuang, M., Huang, L. et al. Lint percentage and boll weight QTLs in three excellent upland cotton (Gossypium hirsutum): ZR014121, CCRI60, and EZ60. BMC Plant Biol 23, 179 (2023). https://doi.org/10.1186/s12870-023-04147-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04147-5