Abstract
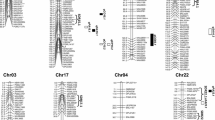
Lint percentage is an important character of cotton yield components and it is also correlated with cotton fibre development. In this study, we used a high lint percentage variety, Baimian1, and a low lint percentage, TM-1 genetic standard for Gossypium hirsutum, as parents to construct a mapping populations in upland cotton (G. hirsutum). A quantitative trait locus/loci (QTL) analysis of lint percentage was performed by using two mapping procedures; composite interval mapping (CIM), inclusive composite interval mapping (ICIM) and the F2:3 populations in 2 years. Six main-effect QTL (M-QTL) for lint percentage (four significant and two suggestive) were detected in both years by CIM, and were located on chr. 3, chr. 19, chr. 26 and chr. 5/chr. 19. Of the six QTL, marker intervals and favourable gene sources of the significant M-QTL, qLP-3(2010) and qLP-3(2011) were consistent. These QTL were also detected by ICIM, and therefore, should preferentially be used for marker-assisted selection (MAS) of lint percentage. Another M-QTL, qLP-19(2010), was detected by two mapping procedures, and it could also be a candidate for MAS. We detected the interaction between two M-QTL and environment, and 11 epistatic QTL (E-QTL) and their interaction with environment by using ICIM. The study also found two EST-SSRs, NAU1187 and NAU1255, linked to M-QTL for lint percentage that could be candidate markers affecting cotton fibre development.


Similar content being viewed by others
References
Abdurakhmonov Y., Buriev Z. T., Saha S., Pepper A. E., Musaev J. A., Almatov A., Shermatov S. E., Kushanov F. N., Mavlonov G. T., Reddy U. K., Yu J. Z., Jenkins J. N., Kohel R. J. and Abdukarimov A. 2007 Microsatellite markers associated with lint percentage trait in cotton, Gossypium hirsutum. Euphytica 156, 141–156.
An C. F., Jenkins J. N., Wu J. X., Guo Y. F. and McCarty J. C. 2010 Use of fibre and fuzz mutants to detect QTL for yield components, seed, and fibre traits of Upland cotton. Euphytica 172, 21–34.
Basra A. S. and Malik C. P. 1984 Development of the cotton fibre. Int. Rev. Cytol. 89, 65–113.
Churchill G. A. and Doerge R. W. 1994 Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971.
Fedak G. 1999 Molecular aids for integration of alien chromatin through wide crosses. Genome 42, 584–591.
Godoy A. S. and Palomo G. A. 1999 Genetic analysis of earliness in Upland cotton (Gossypium hirsutum L.). II. Yield and lint percentage. Euphytica 105, 161–166.
Graves D. A. and Stewart J. M. 1988 Chronology of the differentiation of cotton (Gossypium hirsutum L.) fibre cells. Planta 175, 254–258.
Guo W. Z., Zhang T. Z., Ding Y. Z., Zhu Y. C., Shen X. L. and Zhu X. F. 2005 Molecular marker assisted selection and pyramiding of two QTL for fibre strength in Upland cotton. Acta Genet. Sin. 32, 1275–1285.
Guo W. Z., Cai C. P., Wang C. B., Han Z. G., Song X. L., Wang K. et al. 2007 A microsatellite-based, gene-rich linkage map reveals genome structure, function, and evolution in Gossypium. Genetics 176, 527–541.
Gupta P. K., Varshney R. K. and Sharma P. C. 1999 Molecular markers and their application in wheat breeding. Plant Breed. 112, 369–390.
Han X. M. and Liu Y. X. 2002 Genetic analysis for yield and its components in Upland cotton. Acta Agron. Sin. 28, 533–536.
Jia F., Sun F. D., Li J. W., Liu A. Y., Shi Y. Z., Gong J. W. et al. 2011 Identification of QTL for boll weight and lint percentage of Upland cotton (Gossypium hirsutum L.) RIL population in multiple environments. Mol. Plant Breed. 9, 318–326.
Jiang F., Zhao J. and Zhou L. 2009 Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in Upland cotton. Sci. China Ser. C: Life Sci. 52, 872–884.
John E. M. and Crow L. J. 1992 Gene expression in cotton (Gossypium hirsutum L.) fibre: Cloning of the mRNAs. Proc. Natl. Acad. Sci. USA 89, 5769–5773.
Kearney T. H. 1912 Lint index and lint percentage in cotton breeding. J. Hered. 7, 25–29.
Kohel R. J., Richmond T. R. and Lewis C. F. 1970 Texas marker-1. Description of a genetic standard for Gossypium hirsutum L. Crop Sci. 10, 670–671.
Lacape J. M., Nguyen T. B., Courtois B., Belot J. L., Giband M., Gourlot J. P. et al. 2005 QTL analysis of cotton fibre quality using multiple G. hirsutum × G. barbadense backcross generations. Crop Sci. 45, 123– 140.
Lander E. S. and Kruglyak L. 1995 Genetic dissection of complex traits guidelines for interpreting and reporting linkage results. Nat. Genet. 11, 241–247.
Li C. Q., Guo W. Z., Ma X. L. and Zhang T. Z. 2008 Tagging and mapping of QTL for yield and its components in Upland cotton (Gossypium hirsutum L.) population with varied lint percentage. Cotton Sci. 20, 163–169.
Li C. Q., Guo W. Z. and Zhang T. Z. 2009a fibre initiation development in Upland cotton (Gossypium hirsutum L.) cultivars varying in lint percentage. Euphytica 165, 223–230.
Li C. Q., Guo W. Z. and Zhang T. Z. 2009b Quantitative inheritance of yield and its components in Upland cotton (Gossypium hirsutum L.) cultivars with varied lint percentage. Acta Agron. Sin. 35, 1990–1999.
Li C. Q., Wang C. B., Dong N., Wang X. Y., Zhao H. H., Richard C. et al 2012 QTL detection for node of first fruiting branch and its height in Upland cotton (Gossypium hirsutum L.) Euphytica 188, 441–451.
Li W. H., Hu X Y., Shen W. W., Song Y. P., Xu J. A. and Li Q. 2000 Genetic analysis of important economic traits in Upland cotton (G. hirsutum L.) Cotton Sci 12, 81–84.
Liu Q. M., Jiang J. H., Niu F. A., He Y. J. and Hong D. L. 2012 QTL analysis for seven quality traits of japonica rice based on three genetic statistical models. Chin. J. Rice Sci. 26, 283–290.
Liu R. Z., Wang B. H., Guo W. Z., Qin Y. S., Wang L. G., Zhang Y. M. and Zhang T. Z. 2011 Quantitative trait loci mapping for yield and its components by using two immortalized populations of a heterotic hybrid in Gossypium hirsutum L. Mol. Breed. 2, 297–311.
McCouch S. R., Cho Y. G., Yano M., Paul E., Blinstrub M., Morishima H. and Kinoshita T. 1997 Report on QTL nomenclature. Rice Genet. News 14, 11–13.
Mei M., Syed N. H., Gao W., Thaxton P. M., Smith C. W., Stelly D. M. and Chen Z. J. 2004 Genetic mapping and QTL analysis of fibre-related traits in cotton (Gossypium L.) Theor. Appl. Genet. 108, 280–291.
Nguyen T. B., Giband M., Brottier P., Risterucci A. M. and Lacape J. M. 2004 Wide coverage of tetraploid cotton genome using newly developed microsatellite markers. Theor. Appl. Genet. 109, 167–175.
Pan J. J. 1998 Cotton breeding. China Agriculture Press, Beijing, China.
Paterson A. H., Brubaker C. L. and Wendel J. F. 1993 A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP and PCR analysis. Plant Mol. Biol. Rep. 11, 112–127.
Qin H. D., Guo W. Z., Zhang Y. M. and Zhang T. Z. 2008 QTL mapping of yield and fibre traits based on a four-way cross population in Gossypium hirsutum L. Theor. Appl. Genet. 117, 883– 894.
Ren Y., He Z. H., Li J., Morten L., Wu L., Bai B. et al. 2012 QTL mapping of adult-plant resistance to stripe rust in a population derived from common wheat cultivars Naxos and Shanghai 3/Catbird. Theor. Appl. Genet. 125, 1211–1221.
Saha S., Jenkins J. N., Wu J., McCarty J. C., Gutierrez O. A., Percy R. G. et al. 2006 Effects of chromosome-specific introgression in Upland cotton on fibre and agronomic traits. Genetics 172, 1927–1938.
Shen X. L., Guo W. Z., Lu Q. X., Yuan Y. L. and Zhang T. Z. 2007 Genetic mapping of quantitative trait loci for fibre quality and yield trait by RIL approach in Upland cotton. Euphytica 155, 371–380.
Su C. F., Lu W. G., Zhao T. J. and Gai J. Y. 2010 Verification and fine mapping of QTL conferring days to flowering in soybean using residual heterozygous lines. Chin. Sci. Bull. 55, 499–508.
Sun F. D., Zhang J. H., Wang S. F., Gong W. K., Shi Y. Z., Liu A. Y. et al. 2012 QTL mapping for fibre quality traits across multiple generations and environments in Upland cotton. Mol. Breed. 30, 569–582.
Ulloa M. and Meredith W. R. 2000 Genetic linkage map and QTL analysis of agronomic and fibre quality traits in an intraspecific population. J. Cotton Sci. 4, 161–170.
Van Ooijen J. W. and Voorrips R. E. 2001 JoinMap R Version 3.0: software for the calculation of genetic linkage maps. CPRO-DLO, Wageningen.
Wan Q., Zhang Z. S., Hu M. C., Chen L., Liu D. J., Chen X. et al. 2007 T1 locus in cotton is the candidate gene affecting lint percentage, fibre quality and spiny bollworm (Earias spp.) resistance. Euphytica 158, 241–247.
Wang F. R., Liu R. Z., Wang L. M., Zhang C. Y., Liu G. D., Liu Q. H. et al. 2007 Molecular markers of Verticillium wilt resistance in Upland cotton (Gossypium hirsutum L.) cultivar and their effects on assisted phenotypic selection. Cotton Sci. 19, 424–430.
Wang J. K., Li H. H., Zhang L. Y. and Meng L. 2012 Users’ Manual of QTL IciMapping Version 3.2. The Quantitative Genetics Group, Institute of Crop Science. Chinese Academy of Agricultural Sciences (CAAS), Beijing, China.
Wang Q. L. 2004 The breeding report for the new cotton variety of Baimian1. J. Henan Vocation-Tech. Teach. Coll. 32, 1–3.
Wang S., Basten C. J. and Zeng Z. B. 2005 Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC.
Wu J. X., Gutierrez O. A., Jenkins J. N., McCarty J. C. and Zhu J. 2009 Quantitative analysis and QTL mapping for agronomic and fibre traits in an RI population of Upland cotton. Euphytica 165, 231–245.
Wu M. Q., Zhang X. L., Nie Y. C. and He D. H. 2003 Localization of QTL for yield and fibre quality traits of tetraploid cotton cultivar. Acta Genet. Sin. 30, 443–452.
Xing G. N., Zhou B., Wang Y. F., Zhao T. J., Yu D. Y., Chen S. Y. and Gai J. Y. 2012 Genetic components and major QTL confer resistance to bean pyralid (Lamprosema indicata Fabricius) under multiple environments in four RIL populations of soybean. Theor. Appl. Genet. 125, 859–875.
Xu C. N., Bai C. Q. and Jia J. Z. 1995 Crop Cultivation and Physiology Science Symposium in China. 76.
Yin J. M., Wu Y. T., Zhang J. and Zhang T. Z. 2002 Tagging and mapping of QTL controlling lint yield and yield components in Upland cotton (Gossypium hirsutum L.) using SSR and RAPD Markers. Chin. J. Biotechnol. 18, 162–166.
Yu J. W., Zhang K, Li S. Y., Yu S. X., Zhai H. H, Wu M. et al. 2013 Mapping quantitative trait loci for lint yield and fibre quality across environments in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Theor. Appl. Genet. 126, 275–287.
Zhang D. G., Kong F. L., Zhang Q. Y., Liu W. X., Yang F. X., Xu N. Y. et al. 2003a Genetic improvement of cotton varieties in the Yangtse Valley in China since 1950s. Improvement on yield and yield components. Acta Agron. Sin. 29, 208–215.
Zhang J., Guo W. Z. and Zhang T. Z. 2002 Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor. Appl. Genet. 105, 1166–1174.
Zhang T. Z., Yuan Y. L., Yu J., Guo W. Z. and Kohel R. J. 2003b Molecular tagging of a major QTL for fibre strength in Upland cotton and its marker-assisted selection. Theor. Appl. Genet. 106, 262–268.
Zhang X. L., Gao J. G., Song G. L., Liu F. F., Li S. H., Liu K. F. et al. 2009 Additive and epistatic effects QTL analysis on Upland cotton CRI-G6. Mol. Plant Breed. 2, 312–320.
Zhang Z. S., Xiao Y. H., Luo M., Li X. B., Luo X. Y., Hou L. et al. 2005 Construction of a genetic linkage map and QTL analysis of fibre related traits in Upland cotton (Gossypium hirsutum L.) Euphytica 144, 91–99.
Acknowledgements
This work was supported by Grants from High-tech Program 863 (2012AA101108), National Natural Science Foundation (31371677), National Major Project of Transgenic Breeding (2012ZX08013007-001-004), Natural Science Research Project of Henan Province (2010A210006).
Author information
Authors and Affiliations
Corresponding author
Additional information
[Wang M., Li C. and Wang Q. 2014 Quantitative trait loci mapping and genetic dissection for lint percentage in upland cotton (Gossypium hirsutum). J. Genet. 93, xx–xx]
Rights and permissions
About this article
Cite this article
WANG, M., LI, C. & WANG, Q. Quantitative trait loci mapping and genetic dissection for lint percentage in upland cotton (Gossypium hirsutum). J Genet 93, 371–378 (2014). https://doi.org/10.1007/s12041-014-0385-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-014-0385-9