Abstract
Background
Mammalian gonadal development is crucial for fertility. Sexual differentiation, meiosis and gametogenesis are critical events in the process of gonadal development. Abnormalities in any of these events may cause infertility. However, owing to the complexity of these developmental events, the underlying molecular mechanisms are not fully understood and require further research.
Results
In this study, we employed RNA sequencing to examine transcriptome profiles of murine female and male gonads at crucial stages of these developmental events. By bioinformatics analysis, we identified a group of candidate genes that may participate in sexual differentiation, including Erbb3, Erbb4, and Prkg2. One hundred and two and 134 candidate genes that may be important for female and male gonadal development, respectively, were screened by analyzing the global gene expression patterns of developing female and male gonads. Weighted gene co-expression network analysis was performed on developing female gonads, and we identified a gene co-expression module related to meiosis. By alternative splicing analysis, we found that cassette-type exon and alternative start sites were the main forms of alternative splicing in developing gonads. A considerable portion of differentially expressed and alternatively spliced genes were involved in meiosis.
Conclusion
Taken together, our findings have enriched the gonadal transcriptome database and provided novel candidate genes and avenues to research the molecular mechanisms of sexual differentiation, meiosis, and gametogenesis.
Similar content being viewed by others
Introduction
Infertility is estimated to affect as many as 48.5 million couples worldwide [1]. Its incidence may be as high as 15%. Following cancer, cardiovascular and cerebrovascular diseases, infertility has become the third major disorder that seriously affects human health and causes considerable stress to patients and their families.
Gonads are the important reproductive organs for mammals. The process of gonadal development includes critical developmental events, such as sexual differentiation, meiosis and gametogenesis. Abnormalities in any of these events may cause infertility. The molecular regulatory mechanisms of these crucial developmental events have been the focus of many studies in reproductive biology.
Many genes that are important for gonadal development have been discovered. For example, the Sry, Sox9, M33, Dax1, Wt1, Rspo1, Sf1, Dmrt1, Atrx, Wnt4, Amh, Fgf9, Lhr, Dhh, Insl3, Foxl2 and Sox3 genes play important roles in sexual determination and differentiation [2,3,4,5]. The Stra8, Sycp1, Sycp2, Sycp3, Spo11, Rce8, Dmc1, Dazl, Mlh1 and Msh5 genes regulate meiosis [5,6,7,8,9]. The Figla, Lhx8, Nobox, Sohlh1, Sohlh2, Bax, Ahr, Gdf9, Pten, Scf, Bcl2 and Rps6 genes have roles in follicular and oocyte development [5, 10]. The Adamts2, Bcl2l2, Cadm1, Ddx25, Piwil1, Prm1, Prm2, Tbpl1, Tlp, Tnp1, Tnp2 and Ube2b genes are involved in regulating spermatogenesis [5].
Many previous studies of gonadal development used gene knockout mouse models, and only a small number of genes could be studied at a time. Gene chips were also used to research gonadal development [11,12,13,14]. Although higher throughput was obtained by gene chip analysis, this research was typically limited to the more highly expressed known genes. Classical gene chip analysis was unable to research unknown or low abundance gene transcripts, nor alternative splicing.
In recent years, high-throughput RNA sequencing (RNA-Seq) overcame the above technical shortcomings of gene chips, and also allowed high-throughput investigation of the whole transcriptome [15]. Previous studies have used RNA-Seq to investigate the development of mouse gonads. However, these studies either focused on single sex gonads at certain developmental stages, or specific cell types in the gonads. Gong et al. investigated the transcriptome profiling of mouse testes at three postnatal ages: 6 days postnatal, 4 weeks old and 10 weeks old, representing infant, juvenile and adult stages, respectively [16]. Pan et al. performed a comparative transcriptomic analysis using RNA-Seq of ovaries isolated from mice aged 1 week and 8 weeks old [17]. McClelland et al. carried out transcriptomic analysis of mouse fetal Leydig cells [18]. Sexual differentiation, meiosis and gametogenesis require the joint participation and interaction of germ and somatic cells in the gonads. Therefore, a better understanding of the mechanisms of sexual differentiation may be obtained by comparing female and male gonadal transcriptomes. It is possible that more insight into the molecular mechanisms of sexual differentiation, meiosis and gametogenesis may be obtained by transcriptomic analysis of murine female and male gonads covering the key stages of developmental corresponding to these critical developmental events. However, such a comparison has not been reported to date.
Here, we systematically compared the transcriptomes of female and male gonads covering the developmental stages corresponding to sexual differentiation, meiosis and gametogenesis. Our study used gonads at 12.5 days post-coitum (dpc), at which time female and male gonads begin to show discernible differences, 13.5 dpc when female germ cells begin to undergo meiosis, and male germ cells begin to stop mitosis, 16.5 dpc when most female germ cells are undergoing meiosis, and male germ cells arrest at the G1/G0 stage, and 6 days post-partum (dpp) when most male germ cells are at the spermatogonial stem cell stage. The current results provide novel candidate genes and potential pathways for further investigation into the molecular mechanisms of sexual differentiation, meiosis and gametogenesis in gonadal development.
Materials and Methods
Animals
Eight-week-old C57BL/6 mice were purchased from the SLAC Laboratory Animal Co., Shanghai, China. Mice were housed in a 12 h light: 12 h dark cycle. Protocols and use of animals for this study were approved by the Institutional Animal Care and Use Committee of Shanghai (SYXK-2018-0028) and were conducted in accordance with the National Research Council Guide for Care and Use of Laboratory Animals.
Gonad Collection and RNA Extraction
Timed matings were performed by placing a male mouse with two females in a cage. Females were checked for the presence of vaginal plugs the next morning. Noon of the day when the mating plug was observed was designated 0.5 dpc. On the relevant days of gestation (12.5, 13.5 and 16.5 dpc), pregnant females were euthanized with carbon dioxide. Embryonic gonads were dissected free of the mesonephros, snap frozen in liquid nitrogen, and stored at − 80 °C. The gonads of 6 dpp mice were also collected, immediately frozen in liquid nitrogen and stored at − 80 °C.
Total RNA was isolated from the gonads of 12.5, 13.5 and 16.5 dpc and 6 dpp mice using TRIzol reagent (Invitrogen, Waltham, Massachusetts, USA) according to the manufacturer’s instructions. An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA) was used to measure total RNA quantity and assess RNA integrity.
RNA-Seq Library Construction and Sequencing
The cDNA library was constructed with a SMARTer® Ultra Low Input RNA for lllumina® Sequencing kit (Clontech Laboratories, Mountain View, USA). Briefly, first-strand cDNA was synthesized using SMARTScribe Reverse Transcriptase and then purified using SPRI Ampure Beads. After amplification using the Polymerase Mix, double-stranded complementary DNA was purified using SPRI Ampure Beads.
High-throughput sequencing was performed by Shanghai Biotechnology Inc. (Shanghai, China) via 100-nt paired-end sequencing on an Illumina HiSeq 2500 system (Illumina, San Diego, USA).
Reads Mapping and Measurement of Gene Expression
After sequencing, clean reads were obtained by removing reads containing the adaptor sequences, reads with > 5% ambiguous bases, and low-quality reads, then mapped to the mouse genome (version: mm10_GRCm38) using TopHat software (Version 2.1.1). Gene expression level was calculated using the fragments per kilobase per million mapped reads method.
Analysis of Differentially Expressed Genes
Analysis of Differentially Expressed Genes (DEGs) was performed by the DESeq package in R language (Version 1.36.0). Genes with a fold change (FC) ≥ 2, and false discovery rate (FDR) < 0.05 were assigned as differentially expressed.
The Series-Cluster analysis of expression profiles of DEGs was performed by using the STEM method (http://www.cs.cmu.edu/~jernst/st/). Significant profiles were identified using the Fisher’s exact test and multiple comparisons.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes
Genes were submitted to the databases of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) for enrichment analysis of the significant GO terms and KEGG pathways. Statistically over-represented GO terms and KEGG pathway categories were obtained by applying a Fisher’s exact p-value cutoff < 0.05 and correcting for multiple testing with the Benjamini-Hochberg false discovery rate. Moreover, we built a pathway-act-network of the enriched pathways according to the relationships identified between the pathways in the KEGG database.
Weighted Gene co-Expression Network Analysis
Weighted gene co-expression network analysis(WGCNA) was used to create a co-expression network using the R package (Version 1.68) according to our previous publication [19]. Briefly, to ensure a scale free topology of the network, an adjacency matrix was established by transforming a pairwise Pearson’s correlation matrix of expression values using a power function. The topological overlap measure (TOM) was calculated using the adjacency matrix, and used to cluster genes with a highly similar co-expression relationship into a network module.
Alternative Splicing
Alternatively spliced transcripts were identified by the software AS detector (ASD) (Version 1.0) using the Fisher’s exact test as described [20]. Alternative splicing events were adjusted using Jensen-Shannon divergence (FDR < 0.05).
Quantitative Real-Time Reverse Transcription PCR
Reverse transcription was performed using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme, Nanjing, China). The quantitative real-time reverse transcription PCR (qPCR) was performed using FastStart Universal SYBR Green Master Mix (Roche) according to the procedure described previously [21]. The 2−ΔΔCt method was used to calculate the relative expression of genes using the ABI 7500 System Software (V2.0.4), and gene expression levels were normalized to the Gapdh level. The primers used for qPCR are shown in Table 1.
Statistical Analysis
For each group, three independent experiments were replicated. The data are expressed as the mean ± SEM. Data were statistically analyzed with ANOVA, followed by the Fisher’s least significant difference test with R software (Version 3.5.1). Differences were considered significant at p < 0.05.
Results
Sex-Biased Gene Expression in Mouse Gonads
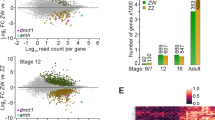
In order to screen candidate genes regulating gonadal sexual differentiation, we initially screened the differentially expressed genes in female and male gonads at the same stage of development. Wayne analysis was then performed to identify the genes with higher expression in all four stages of development, and these genes were defined as sex-biased expressed genes that may participate in regulating gonadal sexual differentiation.
One hundred and thirty-seven male-biased and 187 female-biased expressed genes were obtained (Fig. 1 and Additional file 3 Table. S1, Additional file 4 S2). These genes were classified into 3 categories. The first category included genes functionally annotated and reported to play an important role in sex determination and differentiation, such as Sox9 [22, 23], Xist [24], Amh [25], Insl3 [26], Emx2 [26], Dhh [26], Wnt4 [27, 28], Foxl2 [29] and Fst [26]. The second category included genes with functional annotations but no reported involvement in sex determination and differentiation, including Erbb3, Mt3, Gas6, Lzts1, Lrrn1, Olfm1 and Lgr6. The third category had genes with no functional annotations, such as Gm30587, GM6705, Gm11734 and Gm31399.
Validation of Representative Sex-Biased Gene Expression by qPCR
To validate the screened sex-biased expressed genes, twelve genes (Cst9, Zbtb7c, Lgr6, Gas6, Erbb3, Erbb4, Zfp819, Rec8, Sox8, Sox9, Wnt4 and Foxl2) were randomly selected and their ovarian and testicular expression levels at different stages of development were detected by qPCR. The results showed that Cst9, Erbb3, Erbb4, Zfp819, Sox8 and Sox9 had biased expression in male gonads, whereas Lgr6, Zbtb7c, Gas6, Rec8, Wnt4 and Foxl2 had biased expression in female gonads (Fig. 2). The results of qPCR were consistent with RNA-seq.
Protein–Protein Interaction Network of Mouse Gonadal Sex-Biased Gene Expression
The protein–protein interaction (PPI) network of mouse gonadal sex-biased expressed genes was constructed by using String and Cytoscape software. Twenty nodal genes were connected via more than 10 edges in the network; Wnt4, Dhh, Amh, Bmp2, Fst, Sox9, Spp1, Cd44, Cyp11a1, Cyp17a1, Erbb3, Erbb4, Fshr, Lhcgr, Mapk4, Prkg2, Calb1, Nos1, Acta2 and Cacna1d (Additional file 1 Figure S1 and Tab. 2). There were defined as hub genes in the network, suggesting they may play a key regulatory role in the process of gonadal sexual differentiation.
Among these 20 hub genes, Sox9, Cyp17a1, Cyp11a1, Dhh, Amh, Fshr, Lhcgr, Wnt4, Fst and Bmp2 are well-known sex differentiation genes. The function of the other 10 hub genes with respect to sexual differentiation remains unknown. Interestingly, Erbb3, Erbb4, Prkg2 and Mapk4 encode protein kinases. Protein kinases catalyze protein phosphorylation and transfer the γ-phosphate groups of adenosine triphosphate (usually ATP) to amino acid residues of substrate proteins. The four protein kinase genes suggest that protein phosphorylation may play an important role in sexual differentiation.
Time Series Cluster Analysis of Gonadal Gene Expression during Development
Time series cluster analysis was performed to identify the global trends and model profiles of gonadal gene expression according to signal densities in the 12.5, 13.5 and 16.5 dpc, and 6 dpp temporal sequence. We identified 26 possible gene expression profile patterns (numbered 0–25), which represent the overall expression patterns (Fig. 3a, 4a). Eleven patterns (No. 0, 1, 9, 10, 12, 13, 14, 15, 16, 17 and 25) showed significance (P < 0.05) in female gonadal development, and 10 patterns (No. 0, 3, 9, 12, 13, 14, 15, 16, 23 and 25) showed significance (P < 0.05) in male gonadal development (Fig. 3a, 4a).
Time series cluster analysis of gene expression during female gonadal development. a Differentially expressed genes in the developing female gonads were separated into 26 possible model profiles, including 11 with significant changes in gene expression. b GO enrichment analysis of genes in profile 25 from the female gonad. c KEGG enrichment analysis of genes in profile 25 from the female gonad. d Pathway-Act-Network analysis
Time series cluster analysis of gene expression during male gonadal development. a Differentially expressed genes in the developing male gonads were separated into 26 possible model profiles, including 10 with significant changes in gene expression. b GO enrichment analysis of genes in profile 25 from the male gonad
For both female and male gonads, genes in pattern No. 25 showed increased expression levels during gonadal development, suggesting these genes may be crucial for gonadal development. Significantly enriched GO terms from pattern No. 25 are illustrated in Fig. 3b and 4b. For the female gonads, pattern No. 25 contained 102 genes and significantly enriched GO terms closely correlated with mRNA splicing, positive regulation of the MAPK cascade, N-glycan fucosylation, and negative regulation of oocyte development (Fig. 3b). This list was also significantly enriched for genes associated with the p53 signaling, N-Glycan biosynthesis signaling, and cGMP-PKG signaling pathways (Fig. 3c). To further understand the importance of pathway interactions, and to screen key pathways for significant roles in female gonadal development, we built a Pathway-Act-Network according to the direct or systemic interactions assigned between pathways in the KEGG database (Fig. 3d). Key pathways were identified, including apoptosis signaling and protein processing in endoplasmic reticulum signaling pathways, as shown in Fig. 3d. In addition, we found 9 unannotated ncRNAs in pattern No. 25, designated LOC102639808, LOC102640507, LOC102637464, LOC102637824, LOC102638234, LOC102634191, LOC102635948, LOC102637233 and LOC102631989.
Similarly, for the male gonads, pattern No. 25 contained 134 genes and had significantly enriched GO terms closely correlated with phosphorylation, protein deubiquitination, 9-cis-retinoic acid biosynthesis and sperm chromatin condensation (Fig. 4b). This list also included genes that were significantly enriched in mucin type O-glycan biosynthesis signaling and calcium signaling pathways. Seventeen unannotated ncRNAs were found in this pattern, designated LOC102631654, LOC102631845, LOC102632148, LOC102632349, LOC102634896, LOC102635066, LOC102635096, LOC102636262, LOC102636409, LOC102637414, LOC102637640, LOC102637873, LOC102637942, LOC102639461, LOC102640347, LOC102640483 and LOC102640949.
Weighted Gene Co-Expression Network Analysis
In mouse female gonads, some germ cells were reported to enter meiosis at 13.5 dpc [7, 30], and most germ cells were undergoing meiosis at 16.5 dpc [6, 7]. To study genes that regulate meiosis in germ cells, transcriptome profiles from female gonads were subjected to WGCNA. Using this unsupervised and unbiased analysis, we identified six distinct co-expression modules corresponding to clusters of correlated transcripts (Fig. 5). The co-expression relationship of modules between samples is shown in Fig. 5b. The GO functional enrichment analysis was performed for genes in each module. Genes in the blue module were found to be significantly enriched for meiotic GO items, suggesting that genes in the blue module may be closely associated with meiosis.
In order to obtain core genes that regulate meiosis during gonadal development from 12.5 dpc to 6 dpp, genes significantly enriched for meiosis according to GO analysis of the blue module were selected to construct a gene co-expression network (Additional file 2 Fig. S2). According to co-expression relationships, Mei1, Meiob and Sycp2 were the core genes in the network, suggesting they may have a crucial role in the regulation of meiosis during gonadal development from 12.5 dpc to 6 dpp.
Alternative Splicing
Alternative splicing is regulated during gene expression and results in a single gene encoding multiple mRNAs and proteins. In this process, particular exons of a gene may be included or excluded from the final processed mRNA. Consequently, the proteins translated from alternatively spliced mRNAs may differ in their amino acid sequences and biological functions. Alternative splicing events of transcripts during mouse gonadal development and sexual differentiation were predicted by software AS detector; 1902 and 1559alternative splicing events were identified during female and male gonadal development, respectively, and 962 alternative splicing events were identified during gonadal sexual differentiation. Cassette exons and alternate start sites were the main forms of alternative splicing during murine gonadal development and sexual differentiation (Fig. 6).
Composition of eight types of alternative splicing (AS) patterns in mouse gonads at different stages of developmental stages and during sexual differentiation. a Composition of eight AS patterns in female mouse gonads at different developmental stages. b Composition of eight AS patterns in male mouse gonads at different developmental stages. c Composition of eight AS patterns in gonads during sexual differentiation
It is noteworthy that GO functional enrichment analysis of the differentially-expressed and alternatively spliced genes found that a considerable number were involved in meiosis (Additional file 5 Tab. S3). For example, Dazl was found to exhibit cassette-type alternative splicing at exon 9. Dmc1 was shown to have cassette-type alternative splicing at exons 6, 8 and 11, and multiple cassette-type alternative splicing at exons 7 and 9. These results suggest that alternative splicing may be involved in regulating meiosis.
Dynamic Expression Patterns of miRNAs Involved in Gametogenesis
Distinct miRNAs contribute to gametogenesis [31, 32]. The abnormal expression of specific miRNAs is associated with certain human reproductive dysfunctions. The expression level of such miRNAs can be used as molecular biomarkers to diagnose infertility [33]. In order to study the dynamic expression pattern of miRNAs involved in gametogenesis, we entered “miRNA”, “oogenesis”, “spermatogenesis”, “ovary”, and “testis” keywords into PubMed literature searches. Some miRNAs were previously reported to regulate gametogenesis [34, 35]. We then analyzed the expression of these miRNAs in the gonads of both sexes at embryonic stage (12.5, 13.5 and 16.5 dpc) and infant stage (6 dpp), as shown in Fig. 7.
Most of the miRNAs were expressed in both embryonic and infant stage gonads. The dynamic expression patterns of miR-143, miR-145, miR-214, Lin28 and miR-320 were consistent in the gonads of both sexes. Among them, the expression levels of mir-143, mir-145 and mir-214 generally increased with gonadal development. The expression patterns of miR-130b, miR-17-92, miR-106b, miR-196a and miR-135a showed sexual dimorphism, and mir-135a exhibited female-biased expression.
Discussion
Although the testes and ovaries have different structures and functions, they develop from a common sexually undifferentiated bipotential anlage [36]. Sexual differentiation is essential for gonadal development and function. However, owing to the complexity of the event, the underlying molecular mechanisms remain to be fully understood and require further research. We screened 137 male-biased and 187 female-biased expressed genes, which may be important candidates for regulating gonadal sexual differentiation.
Some classic genes known to play an important role in sexual differentiation were found in the identified genes of this study, such as Sox9, Xist, Foxl2, Wnt4, Amh, Insl3, Dhh, Emx2 and Fst, indicating that our method for screening target genes was reliable. For instance, Sox9 is known to be highly expressed in pre-Sertoli cells of male gonads, and plays a key role in male sex determination by the upregulation of genes associated with testicular differentiation (e.g. Amh, Fgf9, Dhh and Pdgf) and downregulation of genes associated with ovarian development (e.g. Wnt4 and Ctnnb1) [37]. Homozygous loss of Sox9 in XY mice leads to ovarian development and Sox9 overexpression in XX mice leads to testicular development [23]. Mishina et al. reported that Sertoli cells of the fetal testis produce anti-Müllerian hormone (AMH) that causes the regression of the Müllerian ducts, the anlagen of female reproductive organs (uterus, oviducts, and upper portion of the vagina) [38]. The expression of Wnt4 is required for female sexual differentiation. In XX individuals, activation of the β-catenin signaling pathway by secreted RSPO1 and WNT4 proteins is required for granulosa cell differentiation and functional ovarian development [27].
The sex-biased gene expression that we identified included some transcripts without functional annotation with regard to sexual differentiation, as well as transcripts lacking any functional annotation. The current study has provided a large number of candidate genes and the opportunity to identify novel transcripts regulating gonadal sexual development. For example, Lgr6 exhibited female-biased expression, suggesting it may play a role in female sexual differentiation. Lgr6 encodes a receptor for R-spondins (RSPO1, RSPO2, RSPO3 or RSPO4), and it associates with phosphorylated LRP6 and frizzled receptors that are activated by extracellular Wnt receptors, triggering the canonical Wnt signaling pathway to increase expression of target genes. Kawasaki et al. reported that Lgr6 is involved in tooth development [39], but it remains unknown if Lgr6 plays a role in sexual differentiation.
We identified a group of genes with expression levels that continued to rise during female or male gonadal development, suggesting that these genes may be crucial for gonadal development. A significant proportion of these genes were ncRNAs that have not yet been functionally annotated, such as LOC102639808, LOC102640949, LOC102640507, and LOC102637464. This provides a valuable opportunity to identify previously unknown genes that regulate gonadal development. Our future work will investigate the function of these ncRNAs in the regulation of gonadal development. In addition, candidate genes that may be crucial for the development of female gonads were subjected to GO and KEGG pathway enrichment analysis. We identified GO terms and signaling pathways associated with protein glycosylation, suggesting this modification may be involved in the regulation of female gonadal development. Our future studies will target the role of protein glycosylation in the development of female gonads.
Meiosis is the unique division of reproductive cells that produces haploid gametes. The molecular mechanisms regulating meiosis are very complex and have not been fully elucidated. In the gonads of female mice, germ cells were reported to first enter the meiotic cell cycle at 13.5 dpc [7, 30], with most germ cells undergoing meiosis at 16.5 dpc [6, 7]. Transcriptome profiles of female gonads provide valuable data sources for studying the molecular mechanisms of meiosis. Therefore, WGCNA was used to analyze the female gonadal transcriptome data, revealing a co-expression module of genes related to meiosis. A co-expression network was constructed and three hub genes (Mei1, Meiob and Sycp2) were identified that may play a core regulatory role in meiosis during gonadal development from 13.5 dpc to 6 dpp. Reinholdt et al. reported that Mei1 was required for vertebrate meiosis, and was positioned upstream of Dmc1 in the genetic pathway that operated during mammalian meiosis [40]. Luo et al. reported that MEIOB exhibited single-stranded DNA-binding and exonuclease activities and was essential for meiotic recombination in both sexes [41]. MEIOB was found to be a meiosis-specific paralogue of RPA1, and co-localized with RPA in foci on meiotic chromosomes. MEIOB formed a complex with RPA and SPATA22. The chromatin localization and stability of MEIOB depended on SPATA22, and vice versa [41]. Meiob-null mice exhibited meiotic failure and sterility of both sexes [41]. During meiosis, the arrangement of homologous chromosomes is tightly regulated by the synaptonemal complex [42]. Each synaptonemal complex consists of two axial/lateral elements, and numerous transverse filaments [42]. Yang et al. reported that SYCP2 was a primary determinant of axial/lateral elements and was required for the incorporation of SYCP3 into synaptonemal complexes [42]. The fertility of homozygous Sycp2 mutant mice was sexually dimorphic; males were sterile because of a block in meiosis, whereas females were subfertile with reduced litter sizes [42].
Alternative splicing events for gene transcripts during murine gonadal development and sexual differentiation were predicted by the software AS detector. Interestingly, a considerable portion of differentially expressed and alternatively spliced genes were associated with meiosis, suggesting that meiosis may be regulated by alternative splicing. Substantial evidence suggests that pre-mRNA splicing is an important regulator of mouse spermatogenesis [43]. The stage-enriched expression of splicing proteins and substantial changes in alternative splicing patterns around meiosis suggest that alternative splicing may be critical for the mitotic-to-meiotic transition during mouse spermatogenesis [43]. Liu et al. reported that male mice were sterile after Bcas2, an alternative splicing regulator, was knocked out in spermatogonial stem cells [43]. Although the spermatogonia were grossly normal, spermatocytes in meiotic prophase I were scarce and meiotic events were absent in the BCAS2-depleted testes [43]. Moreover, these results suggested that BCAS2 may be involved in the splicing of Dazl, an intrinsic germ cell factor that promotes the initiation of meiosis [43]. However, the regulation of alternative splicing and its role in meiosis may be very complex, and the current research may be the tip of the iceberg.
It is known that miRNA plays an important regulatory role in gametogenesis [31, 32]. Polycystic ovary syndrome, premature ovarian failure and other reproductive disorders are associated with the abnormal expression of specific miRNAs. Selected miRNAs, which contain about 22–25 nucleotides, are often used as a molecular diagnostic markers for infertility owing to the advantages of the small molecular size [33]. When screening for sex-biased gene expression, we found that mir-135a showed female-biased expression, suggesting that it may be involved in regulating female sexual differentiation. Moritoki et al. reported that miR-135a contributed to the maintenance of spermatogonial stem cells by regulating FoxO1 [44]. However, there have been no reports on its involvement in sexual differentiation.
We studied the dynamic expression changes of miRNAs associated with gametogenesis in embryonic and early postnatal gonads, and found that the expression level of mir-196a was higher in male compared with female gonads. The expression level of mir-196a showed an upward trend during the development of male gonads, suggesting it may be important for gonadal development. Rah et al. reported that a putative gene-gene interaction between miR-146 and miR-196a2 may be involved in the development of premature ovarian failure [45]. However, whether this interaction is involved in the regulation of male gonadal development remains to be studied. It is noteworthy that during female gonadal development, the expression of all six members of the miR-17-92 cluster (miR-17, miR-18, miR-19a, miR-20a, miR-19b and miR-92a), was significantly higher at 16.5 dpc than that at other developmental periods. This is a critical stage of female germ cell development, suggesting that mir-17-92 may be involved in regulating oogenesis. In agreement with this finding, our past research showed that ablation of the mir-17-92 cluster in germ cells caused subfertility in female mice, and miR-17-92 participated in the regulation of oogenesis [21].
In this study, the murine transcriptomes of female and male gonads were systematically compared during stages of development corresponding to sexual differentiation, meiosis and gametogenesis. We obtained good quality and reliable transcriptome data from gonads during the critical periods of the above developmental stages, enriching the overall mouse gonadal transcriptome data. Some unannotated candidate genes that may participate in the regulation of sexual differentiation and development were also identified. Our current findings provide novel candidate genes and avenues for further research into the molecular mechanisms of sexual differentiation, meiosis, gametogenesis and other key developmental events in gonadal development.
Conclusions
Our findings have enriched the gonadal transcriptome database and provided novel candidate genes and avenues to research the molecular mechanisms of sexual differentiation, meiosis, gametogenesis, and other key developmental events in gonadal development.
Availability of Data and Materials
Sequence data were submitted to the GEO database under accession number GSE117590.
Abbreviations
- AMH:
-
Anti-Müllerian hormone
- ASD:
-
Software AS detector
- DEGs:
-
Differentially Expressed Genes
- Dpc:
-
Days post-coitum
- Dpp:
-
Days post-partum
- FDR:
-
False discovery rate
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- PPI:
-
The protein–protein interaction
- RNA-Seq:
-
High-throughput RNA sequencing
- TOM:
-
The topological overlap measure
- WGCNA:
-
Weighted gene co-expression network analysis
References
Kitchen H, Aldhouse N, Trigg A, Palencia R, Mitchell S. A review of patient-reported outcome measures to assess female infertility-related quality of life. Health Qual Life Outcomes. 2017;15(1):86.
Makiyan Z. Studies of gonadal sex differentiation. Organogenesis. 2016;12(1):42–51.
Windley SP, Wilhelm D. Signaling pathways involved in mammalian sex determination and gonad development. Sex Dev. 2015;9(6):297–315.
Quinn A, Koopman P. The molecular genetics of sex determination and sex reversal in mammals. Semin Reprod Med. 2012;30(5):351–63.
Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–213.
Spiller CM, Bowles J, Koopman P. Regulation of germ cell meiosis in the fetal ovary. Int J Dev Biol. 2012;56(10–12):779–87.
Feng CW, Bowles J, Koopman P. Control of mammalian germ cell entry into meiosis. Mol Cell Endocrinol. 2014;382(1):488–97.
Inagaki A, Schoenmakers S, Baarends WM. DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics. 2010;5(4):255–66.
Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet. 2010;11(2):124–36.
Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr Rev. 2009;30(6):624–712.
Piprek RP. Methods for the study of gonadal development. Results Probl Cell Differ. 2016;58:383–426.
Combes AN, Bowles J, Feng CW, Chiu HS, Khoo PL, Jackson A, Little MH, Tam PP, Koopman P. Expression and functional analysis of Dkk1 during early gonadal development. Sex Dev. 2011;5(3):124–30.
Coveney D, Cool J, Oliver T, Capel B. Four-dimensional analysis of vascularization during primary development of an organ, the gonad. Proc Natl Acad Sci U S A. 2008;105(20):7212–7.
Nicol B, Yao HH. Gonadal identity in the absence of pro-testis factor SOX9 and pro-ovary factor Beta-catenin in mice. Biol Reprod. 2015;93(2):35.
Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–41.
Gong W, Pan L, Lin Q, Zhou Y, Xin C, Yu X, Cui P, Hu S, Yu J. Transcriptome profiling of the developing postnatal mouse testis using next-generation sequencing. Sci China Life Sci. 2013;56(1):1–12.
Pan L, Gong W, Zhou Y, Li X, Yu J, Hu S. A comprehensive transcriptomic analysis of infant and adult mouse ovary. Genomics Proteomics Bioinformatics. 2014;12(5):239–48.
McClelland KS, Bell K, Larney C, Harley VR, Sinclair AH, Oshlack A, Koopman P, Bowles J. Purification and transcriptomic analysis of mouse fetal Leydig cells reveals candidate genes for specification of gonadal steroidogenic cells. Biol Reprod. 2015;92(6):145.
Wu C, Xu B, Li X, Ma W, Zhang P, Chen X, Wu J. Tracing and characterizing the development of transplanted female germline stem cells in vivo. Mol Ther. 2017;25(6):1408–19.
Zhou X, Wu W, Li H, Cheng Y, Wei N, Zong J, Feng X, Xie Z, Chen D, Manley JL, et al. Transcriptome analysis of alternative splicing events regulated by SRSF10 reveals position-dependent splicing modulation. Nucleic Acids Res. 2014;42(6):4019–30.
Wang J, Xu B, Tian GG, Sun T, Wu J. Ablation of the MiR-17-92 MicroRNA cluster in germ cells causes subfertility in female mice. Cell Physiol Biochem. 2018;45(2):491–504.
She ZY, Yang WX. Sry and SoxE genes: how they participate in mammalian sex determination and gonadal development? Semin Cell Dev Biol. 2017;63:13–22.
Jiang T, Hou CC, She ZY, Yang WX. The SOX gene family: function and regulation in testis determination and male fertility maintenance. Mol Biol Rep. 2013;40(3):2187–94.
Jeong KS, Park JH, Lee S. The analysis of X-chromosome inactivation-related gene expression from single mouse embryo with sex-determination. Biochem Biophys Res Commun. 2005;333(3):803–7.
Carre GA, Greenfield A. The gonadal supporting cell lineage and mammalian sex determination: the differentiation of Sertoli and granulosa cells. Results Probl Cell Differ. 2016;58:47–66.
Biason-Lauber A. Control of sex development. Best Pract Res Clin Endocrinol Metab. 2010;24(2):163–86.
Chassot AA, Gillot I, Chaboissier MC. R-spondin1, WNT4, and the CTNNB1 signaling pathway: strict control over ovarian differentiation. Reproduction. 2014;148(6):R97–110.
Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 2006;4(6):e187.
Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, Treier AC, Treier M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131(4):933–42.
Arora R, Abby E, Ross AD, Cantu AV, Kissner MD, Castro V, Ho HH, Livera G, Laird DJ. Meiotic onset is reliant on spatial distribution but independent of germ cell number in the mouse ovary. J Cell Sci. 2016.
Maalouf SW, Liu WS, Pate JL. MicroRNA in ovarian function. Cell Tissue Res. 2016;363(1):7–18.
Ran M, Chen B, Yin J, Yang A, Jiang M. Advances in miRNA research related to testis development and spermatogenesis. Yi Chuan. 2014;36(7):646–54.
Khazaie Y, Nasr Esfahani MH. MicroRNA and male infertility: a potential for diagnosis. Int J Fertil Steril. 2014;8(2):113–8.
Chen X, Li X, Guo J, Zhang P, Zeng W. The roles of microRNAs in regulation of mammalian spermatogenesis. J Anim Sci Biotechnol. 2017;8:35.
Wang L, Xu C. Role of microRNAs in mammalian spermatogenesis and testicular germ cell tumors. Reproduction. 2015;149(3):R127–37.
Piprek RP, Kloc M, Kubiak JZ. Early development of the gonads: origin and differentiation of the somatic cells of the genital ridges. Results Probl Cell Differ. 2016;58:1–22.
Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Dev Biol. 2004;274(2):271–9.
Mishina Y, Rey R, Finegold MJ, Matzuk MM, Josso N, Cate RL, Behringer RR. Genetic analysis of the Mullerian-inhibiting substance signal transduction pathway in mammalian sexual differentiation. Genes Dev. 1996;10(20):2577–87.
Kawasaki M, Porntaveetus T, Kawasaki K, Oommen S, Otsuka-Tanaka Y, Hishinuma M, Nomoto T, Maeda T, Takubo K, Suda T, et al. R-spondins/Lgrs expression in tooth development. Dev Dyn. 2014;243(6):844–51.
Reinholdt LG, Schimenti JC. Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma. 2005;114(2):127–34.
Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun. 2013;4:2788.
Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173(4):497–507.
Liu W, Wang F, Xu Q, Shi J, Zhang X, Lu X, Zhao ZA, Gao Z, Ma H, Duan E, et al. BCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosis. Nat Commun. 2017;8:14182.
Moritoki Y, Hayashi Y, Mizuno K, Kamisawa H, Nishio H, Kurokawa S, Ugawa S, Kojima Y, Kohri K. Expression profiling of microRNA in cryptorchid testes: miR-135a contributes to the maintenance of spermatogonial stem cells by regulating FoxO1. J Urol. 2014;191(4):1174–80.
Rah H, Jeon YJ, Shim SH, Cha SH, Choi DH, Kwon H, Kim JH, Shin JE, Kim NK. Association of miR-146aC>G, miR-196a2T>C, and miR-499A>G polymorphisms with risk of premature ovarian failure in Korean women. Reprod Sci. 2013;20(1):60–8.
Acknowledgements
Not applicable.
Competing of Interests
The authors declare that they have no competing interest.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFC1003501, 2017YFA0504201) and the National Nature Science Foundation of China (81720108017).
Author information
Authors and Affiliations
Contributions
J. W., G.G.T. and Z.Z. conducted all the major experiments, data analysis and wrote the manuscript; B.L. was responsible for qPCR analysis; Q.X. conceived this project; J.W. initiated and supervised the entire project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
Protocols and use of animals for this study were approved by the Institutional Animal Care and Use Committee of Shanghai (SYXK-2018-0028) and were conducted in accordance with the National Research Council Guide for Care and Use of Laboratory Animals.
Consent for Publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Protein–protein interaction network for genes with sex-biased gonadal expression in mice. (PDF 203 kb)
Additional file 2: Figure S2.
Gene co-expression network of meiosis-related genes in the blue module. (PDF 478 kb)
Additional file 3: Table S1.
The list of male-biased expressed genes. (DOCX 15 kb)
Additional file 4: Table S2.
The list of female-biased expressed genes. (DOCX 16 kb)
Additional file 5: Table S3.
A list of differential alternative splicing meiotic genes. (DOC 66 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, J., Tian, G.G., Zheng, Z. et al. Comprehensive Transcriptomic Analysis of Mouse Gonadal Development Involving Sexual Differentiation, Meiosis and Gametogenesis. Biol Proced Online 21, 20 (2019). https://doi.org/10.1186/s12575-019-0108-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12575-019-0108-y