Abstract
Multiple haplotypes at the same rice blast R-gene locus share extremely high sequence similarity, which makes the gene diagnostic method using molecular markers less effective in differentiation from one another. The composition and distribution pattern of deployed R genes/haplotypes in elite rice varieties has not been extensively analyzed. In this study, we employed PCR amplification and sequencing approach for the diagnosis of R-gene haplotypes in 54 Chinese elite rice varieties. A varied number of functional and nonfunctional haplotypes of 4 target major R-gene loci, i.e., Pi2/9, Pi5, Pik, and Pib, were deduced by referring to the reference sequences of known R genes. Functional haplotypes accounted for relatively low frequencies for the Pi2/9 (15%) and Pik (9%) loci but for relatively high frequencies for the Pi5 (50%) and Pib (54%) loci. Intriguingly, significant frequencies of 33%, 39%, 46% of non-functional haplotypes at the Pi2/9, Pik, and Pib loci, respectively, with traceable original donors were identified, suggesting that they were most likely unintentionally spread by using undesirable donors in various breeding programs. In the case of Pi5 locus, only a single haplotype, i.e., Pi5 was identified. The reactions of 54 rice varieties to the differential isolates were evaluated, which showed a good correlation to the frequency of cognate avirulence (Avr) genes or haplotypes in the differential isolates. Four R genes, i.e., Pi2, Piz-t, Pi50, and Pikm were found to contribute significantly to the resistance of the elite rice varieties. Other two genes, Pi9 and Pikh, which were not utilized in rice varieties, showed promising values in breeding durable resistance due to their high resistance frequencies to the contemporary rice blast population. The sequence-based molecular diagnosis provided a promising approach for the identification and verification of haplotypes in different R-gene loci and effective R genes valuable for breeding durable rice resistance to rice blast.
Similar content being viewed by others
Background
Rice is a staple diet for nearly half of the global population (Harlan 1998; Skamnioti and Gurr 2009), and its increase in production to meet the ever-increasing population faces many challenges. Breeding high yield rice varieties, especially hybrid rice is one of the promising methods which has made a great contribution to safeguarding the food supply around the world (Tester and Langridge 2010). However, the high-yield hybrid rice is now seriously threatened by specific biotic stresses (blast, brown planthopper, bacterial blight, false smut, and so on) (Khush and Jena 2009; Wang and Valent 2017). Apart from focusing on yield potential, it is essentially important in improving the adaptability, resistance to biotic and abiotic stresses of rice.
Rice blast, caused by the fungal pathogen Magnaporthe oryzae (M. oryzae), is the most devastating rice disease and conspicuously reduces rice yield and grain quality (Kush and Jena 2009). The rice and rice blast system belong to a typical gene-for-gene system (Flor 1971), in which the host resistance (R) genes show functional correspondence to their cognate pathogen avirulence (Avr) genes (Orbach et al. 2000; Valent and Khang 2010). The co-evolution and interaction of R and Avr gene raises the possibility of a gene-specific arms race leading to diversification of both R and Avr genes (Dodds et al. 2006). Nine rice blast Avr genes have been cloned to date (Wang et al. 2017). The direct and indirect interaction models between R and Avr proteins were illustrated (Li et al. 2009; Yoshida et al. 2009; Kanzaki et al. 2012; Wu et al. 2015; Zhang et al. 2015; Ray et al. 2016). Field efficacy of any R gene in rice varieties is proposed to depend on the frequency of its cognate Avr gene in the rice blast pathogen population, which provides a basis of Avr-gene based diagnosis for the deduction of effectiveness of R genes (Selisana et al. 2017; Olukayode et al. 2019).
On the other hand, over 105 major rice blast R genes have been identified and 32 of them have been molecularly characterized (Li et al. 2017; Wang et al. 2017; Zhao et al. 2018). Many R genes are clustered at the same locus and allelic to each other. For instance, Pi2, Piz-t, Pi9 and Pi50 are allelic at the Pi2/9 locus (Qu et al. 2006; Zhou et al. 2006; Su et al. 2015). Pik, Pikp, Pikm, Pikh, Pi1 and Pi7 are allelic at the Pik locus (Ashikawa et al. 2008; Yuan et al. 2011; Zhai et al. 2011; Hua et al. 2012). Pish, Pi35 and Pi37 are allelic at Pish locus (Lin et al. 2007; Takahashi et al. 2010; Fukuoka et al. 2014). Moreover, a limited number of sequence differences between these R-gene alleles were found to determine their specificities against distinct sets of rice blast isolates (Zhou et al. 2006; Fukuoka et al. 2014; Su et al. 2015). It was also found that functional and non-functional R-gene haplotypes at the same locus from resistant and susceptible rice varieties were distinguished by a few sequence changes (Bryan et al. 2000; Su et al. 2015). The fact that non-functional and functional R-gene haplotypes with distinct resistance specificities embedded in the same locus raises a reasonable concern about the sensitivity and reliability of the use of R-gene specific or linked markers in the diagnosis of the respective R genes present in novel germplasm. In this study, we employ a method combining PCR amplification and subsequent amplicon sequencing of portions of known rice blast R genes determining their function and resistance specificities for the diagnosis of R-gene haplotypes present in elite rice varieties. The function and the resistance specificities of R-gene haplotypes in these varieties are to be further validated using differential rice blast isolates with the reference of the presence of Avr genes.
Methods
Plant Materials
A total of 54 rice accessions were collected in this study. Forty of them were requested from the rice germplasm bank of Hunan Academy of Agricultural Sciences and the other 14 accessions were bought from the seed market. The detail information including name, promotion area and year, female and male parents for all these 54 rice accessions were listed in Additional file 1: Table S1. Eleven IRBLs, IRBLb-B, IRBLks-F5, IRBLkp-K60, IRBLkh-K3, IRBLi-F5, IRBL5-M, IRBLz5-CA, IRBLzt-T, IRBL9-W, IRBLz-Fu and IRBL1-CL, and the rice variety CO39 and NIL-e1 (monogenic line of Pi50) (Su et al. 2015) were also used in this study. Plants were grown in greenhouses at Hunan Hybrid Rice Research Institute.
Fungal Materials and Disease Evaluation
Twenty seven M. oryzae isolates were used for disease evaluation and Avr gene diagnosis in this study. 26 of them were collected from Guangdong province which can represent the field population of M. oryzae in South China and one was collected from Hubei province, China. The name, collection year and place of these isolates were listed in Additional file 2: Table S2.
Rice seedlings at the 3–4 leaf stage were inoculated using these 27 isolates. The spore suspension with 105 spores/ml was applied to plants using an airbrush connected to a source of compressed air. After inoculation, plants were held in the dark room for 24 h with 95–100% relative humidity and 24 °C. Then, plants were transferred to a greenhouse where the temperature was maintained at 25–28 °C and humidity was keep at 70–90%. Seven days after inoculation, disease symptoms were evaluated using a standard 0–9 scale (IRRI 2002), where 0–1 = highly resistant (HR), 2–3 = resistant (R), 4 = moderately resistant (MR), 5–6 = moderately susceptible (MS), 7 = susceptible (S), and 8–9 = highly susceptible (HS).
Molecular Characterization of Haplotypes of Four R and Five Avr Genes/Locus
Genomic DNAs of both rice varieties and rice blast isolates were extracted using CTAB method and diluted to working concentration for further analysis. For the diagnosis of rice R genes, primers targeting the critical sequence portion capable for differentiating different haplotypes of R-gene loci were designed and optimized. For Avr genes in rice blast, primers targeting the entire genic sequence of each Avr gene were designed and optimized. The amplicons of both R-gene haplotypes and Avr genes were completely sequenced. The names, sequences, position and purpose of all the primers were shown in Additional file 3: Table S3. Polymerase chain reaction (PCR) was performed following the manufacturer’s instructions of the Q5 DNA polymerase (New England Biolabs).
Sequence Data Analysis
The PCR amplification products were send to TSINGKE company (Changsha, China) for sequencing. Multiple alignment of DNA sequences was performed using the software SEQUENCHER (Gene Codes Corporation). The sequences of individual genes were downloaded from National Center for Biotechnology Information (NCBI) and used as reference sequence for alignment.
Results
Establishment of the method for the diagnosis of haplotypes at four rice blast R-gene loci
For the diagnosis of R-gene haplotypes, portions of known R genes that are capable for differentiating different haplotypes were targeted for PCR amplification and sequence analysis. Four R-gene loci, i.e., Pi2/9, Pi5, Pik, and Pib, representing those containing multiple functional and nonfunctional haplotypes in different rice varieties, were selected in this study. For the Pi2/9 locus, an approximately 3.8-kb fragment corresponding to 3′ portion of Nbs4-Pi2 at the Pi2 locus (Genbank accession no.: DQ352453) flanked by Pi2/9-F3/R4 was targeted (Additional file 3: Table S3). It is worth noting that a prescreen step by PCR amplification was conducted using Pi2/9-DF1/DR1 to exclude those lines without positive PCR amplification as described previously (Xiao et al. 2017). For the Pik locus, the region coding for the HMA domain of Pikp-1 (Genbank accession no.: HM035360) flanked by RGA4-F3/R3 was targeted (Additional file 3: Table S3). For the Pi5 locus, a 740-bp fragment corresponding to the 3′ portion of Pi5–2 (Genbank accession no.: EU869186) flanked by 09RL09-F2/R2 was targeted (Additional file 3: Table S3). For the Pib locus, the fragment corresponding to the 3′ portion of Pib (Genbank accession no.: AB013448) flanked by Pib-F4/R7 was targeted (Additional file 3: Table S3).
To verify the efficiency and specificity of PCR amplification of the target regions of different R gene, the Lijiangxintuanheigu (LTH)-derived International Rice Research Institute (IRRI) bred blast resistant lines (IRBLs) containing respective R genes were used for the validation and the derived amplicons were proceeded for sequencing confirmation. As Additional file 4: Figure S1 illustrated, the PCR amplification was strong and specific for those IRBLs. On the contrary, no PCR amplicons were obtained in CO39. These PCR-based markers were also found to effectively differentiate DNA samples from different varieties (Additional file 4: Figure S1). Sequences of the amplicons from IRBLs were confirmed to be identical to the respective known R genes, suggesting that these primers are working for the specific amplification of target portions of known R genes at different loci.
Composition and Distribution of Different R-Gene Haplotypes in Elite Rice Varieties
A set of 14 rice varieties registered as top-50 varieties with large planting areas in 1994 and 40 hybrid rice varieties registered as top-50 hybrid rice varieties with large planting areas in 2015 were selected as representatives of elite rice varieties for the survey of the R genes in this study (Additional file 1: Table S1).
PCR-based analyses revealed that 24, 24, 27, and 42 varieties were resolved with expected amplicons for the Pik, Pi2/9, Pi5 and Pib loci, respectively (Table 1). Amplicon sequencing and further cluster analyses revealed that these amplicons can be grouped into 5, 4, 1, and 2 haplotypes for the Pik, Pi2/9, Pi5, and Pib loci, respectively, indicating that many varieties most likely each contain the same R-gene haplotype at these 4 loci. The composition of the R-gene haplotypes at 4 loci was further identified by sequence comparison as follows:
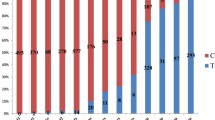
The Pik locus: 5 haplotypes corresponded to Pikm1-LTH (Genbank accession no.: ANO81532), Pik-1 (Genbank accession no.: HM048900), Pi7–1 (Genbank accession no.: HQ660231), Pikm1TS (Genbank accession no.: AB462256), and Pik-1-Shin2 (Genbank accession no.: ADE80950). Out of these 5 Pik haplotypes, Pikm1-LTH and Pik-1-Shin2, are most likely non-functional since their respective varieties LTH and Shin2 were characterized to be susceptible. Surprisingly, Pikm1-LTH was found disproportionally in 20 varieties representative of a frequency of 37% in varieties (Fig. 1 and Table 1). By including Gangyou188 containing Pik-1-Shin2 and another 30 varieties without any Pik haplotypes (Fig. 1 and Table 1), approximately 94% of 54 tested varieties either contain non-functional Pik haplotypes or do not contain any Pik haplotypes. On the contrary, Pik-1, Pikm1-TS, and Pi7–1 were respectively found in 1, 1, and 3 varieties including 2 heterozygous with Pikm1-LTH, representing extremely low frequencies in the elite rice varieties (Table 1 and Additional file 5: Figure S2).
Frequency of different haplotype in 4 R-gene loci. The frequency of alleles was calculated based on percentage of the number of varieties containing the respective R-gene haplotypes in 54 rice varieties. The designation of R-gene haplotypes on the top of each column can be found in Table 1. Different R-gene loci are indicated in different colored boxes
The Pi2/9 locus: 4 haplotypes corresponded Pi2/9-V20, a novel Pi2/9 haplotype (Genbank accession no.: MN630588), Pi2 (Genbank accession no.: DQ352453), Pi50 (Genbank accession no.: KP985761), and Piz-t (Genbank accession no.: DQ352040). Pi2/9-V20 was identified in 18 varieties representative of an approximately 33% of frequency in varieties (Table 1 and Fig. 1). It was further found that Pi2/9-V20 was most likely non-functional since no strong association between its presence and the resistance of the respective variety against differential isolates was identified (Table 1 and Additional file 6: Table S4). Taken together, almost 89% of 54 varieties either contain non-functional Pi2/9 haplotype or do not contain Pi2/9 haplotypes. On the contrary, Pi50, Piz-t, and Pi2 were respectively identified in 1, 1, and 6 varieties including 2 heterozygous with Pi2/9-V20, representing very low frequencies in the elite varieties (Table 1 and Additional file 5: Figure S2).
The Pi5 locus: the single haplotype corresponded to Pi5–2 (Genbank accession no.: EU869186), which was identified in 27 rice varieties representative of a frequency of 50% in elite varieties (Table 1 and Fig. 1).
The Pib locus: 2 haplotypes corresponded to Pib-GII (Genbank accession no.: MN630588) and Pib (Genbank accession no.: AB013448). Pib was identified in 29 varieties representative of an approximately 53.7% of frequency (Table 1 and Fig. 1). Pib-GII, a non-functional haplotype as described previously (Olukayode et al. 2019), was identified in 25 rice varieties (Table 1). It is worth noting that 12 varieties were found to be heterozygous with both Pib and Pib-GII (Table 1).
The Composition and Distribution of Haplotypes of Different Avr Genes in Differential Isolates Used for the Resistance Evaluation of Elite Rice Varieties
A set of 27 M. oryzae isolates likely representative of the contemporary rice blast population was used as differential isolates for the resistance evaluation of 54 rice varieties (Additional file 2: Table S2). It was illustrated that the Avr gene or its haplotype could be used to diagnose the avirulence of the respective isolate to the rice variety containing the cognate R gene (Selisana et al. 2017). We conducted the analysis of the composition and distribution of 5 Avr genes and their haplotypes in these 27 differential isolates detailed as follows:
AvrPii: it was found only in the isolate 98–288 based on the PCR amplification (Table 2). Amplicon sequencing further verified that it was identical to AvrPii (Genbank accession no.: AB498874).
AvrPik: 3 haplotypes corresponded to AvrPik-A, −D, and –E (Genbank accession nos.: AB498875-AB498879). Fourteen isolates contained AvrPik-D (Table 2). Intriguingly, out of these 14 isolates, 12 contained AvrPik-E and 1 contained AvrPik-A, suggesting that most of AvrPik-D containing isolates had an additional AvrPik haplotype. On the contrary, 11 isolates contained only AvrPik-E (Table 2). Taken together, AvrPik-E was identified in 23 isolates, representative of an approximately frequency of 85%. Two isolates were unable to be resolved with any PCR amplicon (Table 2). Pathogenicity of isolates containing AvrPik-D, −E, and -A to Pi1-, Pikh-, and Pikp-containing monogenic lines revealed that AvrPik-E and -A depleted the resistance mediated by Pikp (Additional file 6: Table S4), which is consistent with previous findings (Chaipanya et al. 2017; Selisana et al. 2017).
AvrPi9: It was found in all isolates and amplicon sequencing further revealed that it was identical to AvrPi9 (Genbank accession no.: KM004023) (Table 2).
AvrPiz-t: 3 haplotypes corresponded to AvrPiz-t-H1 ~ AvrPiz-tH3 (Table 2). AvrPiz-t-H1 identical to AvrPiz-t (Genbank accession no.: EU837058) was identified in 19 isolates representative of an approximately 70% of frequency (Table 2). AvrPiz-t-H2 was identified only in the isolate 10–157 and encoded for AvrPiz-tA4IV, caused by a single nucleotide substitution from C to T which corresponded to the 41st residue as described previously (Li et al. 2009). AvrPiz-t-H3 was found in 7 isolates which contained an approximately 6-kb insertion deduced by PCR amplification using a pair of primers in the promoter of AvrPiz-t (AvrPizt-300F/AvrPizt-10R). Amplicon sequencing of this PCR amplicon further revealed that it was an Inago 2-like retrotransposon [98% sequence similarity to the published one (Genbank accession no.: AB334125), which was inserted at the position of -182 bp with a 5-bp “AATGC” target site duplication (Table 2). Besides the insertion of an Inago 2-like element, no sequence difference was found from AvrPiz-t. Both AvrPiz-t-H2 and AvrPiz-t-H3 were deduced to be virulent to Piz-t based on their pathogenicity to Piz-t monogenic line (Additional file 6: Table S4).
AvrPib: 9 different haplotypes designated as AvrPib-H1 ~ AvrPib-H9 were identified based on the sequences of PCR amplicons (Table 2). AvrPib-H1 was identical to AvrPib (Genbank accession no.: KM887844) and identified in only 3 isolates. AvrPib-H2, AvrPib-H4 and AvrPib-H9 were identical to those described previously (Zhang et al. 2015). However, AvrPib-H3 contained a single nucleotide substitution from A to T at the position of + 97 (Genbank accession no.: MN630588), and AvrPib-H5 ~ AvrPib-H8 each contained a Pot3 element at − 56, − 274, − 65, − 125 bp, respectively, based on the AvrPib sequence (Table 2). All the AvrPib haplotypes except AvrPib-H1 were deduced to be virulent to Pib based on their pathogenicity to Pib monogenic line (Additional file 6: Table S4).
Resistance Spectrum Analysis of 54 Rice Varieties to Differential Isolates
The aforementioned 27 differential isolates were used to inoculate all 54 rice varieties at the seedling stage to determine their resistance spectra. A range from 3.7% to 100% of resistance frequencies (RFs) was observed in these 54 rice varieties, indicating that they differed significantly in the resistance to the same rice blast population (Additional file 6: Table S4). Three varieties (Chuanyouhuazhan, Wuyouhuazhan, and Xinrongyouhuazhan) were resistant to all isolates whereas Eyi105 and Ewan were resistant to less than 10% of isolates (Additional file 6: Table S4). It was noted that 23 rice varieties contained multiple known R genes, indicating that R genes were stacked in many varieties (Table 1 and Fig. 2). Among them, 6 varieties contained 3 or 4 R genes and were highly resistant to the differential isolates (Additional file 6: Table S4 and Fig. 2). It is worth noting that 5 of them, i.e., Tianyouhuazhan, Wuyouhuazhan, C-liangyouhuazhan, Xinrongyouhuazhan, and Rongyouhuazhan, were derived from the same male parental line, Huazhan and all contained Pi2, Pi5, and Pib (Additional file 1: Table S1 and Additional file 6: Tables S4). It is thus reasonable to speculate that the restore line Huazhan could contain all or some of these 3 R genes. A relatively positive correlation between resistance and the number of stacked R genes in the varieties was observed based on the regression analysis (Fig. 2), suggesting that the pyramiding of multiple R genes rendered the high level of resistance of varieties to the rice blast isolates.
Resistance frequency of 54 rice varieties. Rice varieties are listed in the order of number of pyramided R genes as in Additional file 6: Table S4. The number of pyramided R genes is indicated by lines. The regression curve is drawn along the resistance frequency
To deduce the contribution of different R genes to the varietal resistance, we included nine monogenic lines including 9 different IRBLs and NIL-e1 for Pi50 in the assay. Different monogenic lines displayed varied RFs ranging from 3.7% to 100% to the differential isolates (Additional file 6: Table S4). The Pi9 monogenic line was resistant to all isolates whereas Pii monogenic line is resistant to only one isolate (Additional file 6: Table S4). RFs of 7 monogenic lines (IRBL9-W, IRBLzt-T, IRBLkh-K3, IRBL1-CL, IRBLkp-K60, IRBLi-F5, and IRBLb-B) evaluated by pathogenicity tests had the same value of the frequency of respective cognate Avr genes in 27 differential isolates (Table 2 and Additional file 6: Table S4). These results reiterated the reliability of the Avr-gene based diagnosis for determining the isolates’ pathotypes to respective R genes (Selisana et al. 2017). It is worth noting that Pikh and Pi1 isogenic lines showed the same RF (93%), which is much higher than the one of Pikp (52%). The discrepancy of the RF between Pikp and Pikh/Pi1 is attributable to the presence of 41% of isolates containing only AvrPik-E which is able to defeat Pikp (Table 2). The result herein verified again the distinct recognition specificities between different Pik and AvrPik alleles (Kanzaki et al. 2012; Selisana et al. 2017). It is noted that the RFs of some monogenic lines were used to infer those of other R-gene alleles i.e., Pikp, Pi1, and Pii for Pi7, Pikm, and Pi5, respectively, since they controlled the same resistance spectra as described previously (Selisana et al. 2017). Based on the RF of each known R gene at the targeted 4 R-gene loci, the contribution rates of known R gene(s) to the varieties were deduced. The interaction pattern of the stacked R genes resulted in distinct effects on the RFs of varieties. For example, the resistance conferred by Pib was perfectly masked by the one controlled by Pi2, leading to no additive effect on RFs of rice varieties containing both of them. On the contrary, no overlapped resistance was identified between Pi7 and Pib, leading to completely additive effects of RFs of rice varieties stacked with both of them (Additional file 6: Table S4).
The Contribution of R Genes to the Resistance of Rice Populations
To evaluate the contribution of R genes to rice resistance at the population level, we calculated the accumulative effects of individual R genes to the resistance of 54 rice varieties against 27 isolates. The total number of resistance reactions attributable to the individual R genes was then summed. The ratio of the summed value against the total reactions was then calculated and represented as the contribution rate of the respective R gene to the rice population. As Fig. 3 illustrated, around 18.9% of accumulative resistance controlled by three R genes, i.e., Pik, Pib, and Pi5, was identified in the population of 1994. However, around 41.7% of resistance conferred by 7 R genes was identified in the population of 2015, indicating that rice varieties in 2015 were overall more resistant than those in 1994. Both Pib and Pi2 contributed more significantly to the resistance than other R genes. Nevertheless, it is worth noting that the larger contribution of Pib to the resistance was partly because of the higher frequency of Pib in rice varieties (Table 1 and Fig. 1). We also measured the percentile of rice varieties resistant to more than 50% of isolates in both populations of 1994 and 2015. It is evident that more varieties in 2015 showed no lower than 50% resistance to the isolates than in 1994 (67.5% versus 50%) (Additional file 7: Figure S3). These data indicated that varieties in 2015 showed a higher level of resistance than those in 1994 to rice blast.
Contribution of different R genes to the resistance of varieties in 1994 and 2015. The percentages of individual R genes contributed to the varietal resistance are indicated in colored boxes corresponding to the different R genes. The accrued percentage from individual R genes is indicated on the top of boxes for the years of 1994 and 2015
We further classified the resistant and susceptible reactions to 4 different categories depending on the presence or absence of known R genes. Category I represented the resistant reaction with the presence of known R gene(s); category II represented the susceptible reaction with the presence of known R gene(s); category III represented the resistant reaction without the presence of known R gene(s); category IV represented the susceptible reaction without the presence of known R gene(s) (Additional file 8: Figure S4). The number of reactions sitting in different categories was summed separately. Around 21.5% of total reactions belonged to category I, demonstrating that the presence of known R gene(s) leads to the varietal resistance against rice blast (Additional file 8: Figure S4). Consistently, around 12.9% of reactions belonging to category IV illustrated another expected outcome in which varieties without known R genes were susceptible. On the contrary, around 26.6% of reactions belonging to category II were susceptible although the varieties contained known R genes, indicating that these R genes eroded in their function already (Additional file 8: Figure S4). Category III represented another outcome in which varieties showed resistance although no known R genes were identified. We postulated that most likely some unknown R genes were attributable to the varietal resistance in this category. It is worth noting that the percentile of category III is much higher than the one of category I (39% versus 21.5%), indicating that unknown R genes rather than known R genes contributed more significantly to the resistance of varieties used in this study (Additional file 8: Figure S4). It was also found that only 14 out of 54 rice varieties in which known R gene(s) contributed not less than 50% of resistance (Additional file 6: Table S4).
Discussions
Resistance to rice blast to a certain level is one of indispensable requirements for varietal releasing in the vetting system in some rice growing countries, such as China (Zeng et al. 2017; Pandian et al. 2018). Therefore, rice blast resistance has been considered as one of the priorities in the rice breeding program. R-gene linked or specific molecular markers were developed and widely used in the mainstream breeding programs via marker aided selection (MAS) (Jiang et al. 2012; Balachiranjeevi et al. 2015; Ni et al. 2015; Ji et al. 2016; Shalini et al. 2016; Wu et al. 2016; Mi et al. 2018). However, many rice blast R-gene haplotypes from different rice lines are almost identical except for a few nucleotide differences, leading to different specificities or even loss of function, e.g, different Pik and Pi2/9 alleles (Zhou et al. 2007; Constanzo and Jia 2010). In this regard, it is critical to develop markers with perfect correlation with their targeted R genes for the diagnosis and selection. A most recent study documented the frequencies of different blast R genes in 161 Indian rice landraces using R-gene linked markers (Yadav et al. 2019). In this study, approximately 12.42% and 85% of landraces contained Pi2 and Piz-t, respectively. Moreover, the landraces containing Piz-t were surprisingly noted to be positive for Piz (Yadav et al. 2019). Contrasting to the extremely high frequency of Pi2/9 alleles described above, only 1 and 5 lines were identified to contain Pi2 and Piz-t out of 1400 rice landraces by referring to the complete sequence of coding region, suggesting a very low frequency of functional Pi2/9 alleles was inherited in rice landraces (Xiao et al. 2017). This contrasting distribution pattern of different alleles at the same R gene locus could be explained to be attributable to different rice population used. Nevertheless, it is necessary to validate the data interpretation by gene sequencing rather than merely by PCR amplification. In this study, we employed a similar method of amplicon sequencing as described previously (Xiao et al. 2017) and applied it for the diagnosis of 4 R-gene loci in a selected set of popular rice varieties particularly hybrids released in the past two decades in China. The fragment amplified and sequenced covered the region mainly determining the specificities of different alleles, capable for distinguishing different alleles from one another. In addition, novel alleles containing sequence variations can also be classified, providing a de novo approach for discovering new variants in addition to diagnosis of known ones. As an example for the haplotypes at the Pi2/9 locus, we identified 1 and 6 rice lines contained Piz-t and Pi2, respectively, suggesting that either Piz-t or Pi2 was not frequently introduced into the elite rice varieties although they were cloned over 10 years ago (Zhou et al. 2006). A similar finding in which the Pi2 gene was not detected in all 88 elite hybrid rice varieties in China using the gene specific marker was reported recently (Wu et al. 2017). In addition to Pi2 and Piz-t, a novel Pi2/9 allele Pi2/9-V20 most likely not responsible for the varietal resistance was identified in a significant number of rice lines, providing a clue for tracing its inheritance in different rice lines. It is worth noting that the current approach is based on the traditional PCR amplification and subsequent sequencing using Sanger method, which is relatively low throughput. Moreover, we note that partial sequence analysis of R-gene haplotypes may not be sufficient to provide accurate diagnosis of blast R genes due to presence of mutations in the un-sequenced regions. However, we would like to insist that comparison of the sequences corresponding to the critical portion of R-gene haplotypes is at least able to differentiate them from known sequences. Furthermore, we focused the analysis in the released varieties rather than in diverse germplasm. A high probability for the deduction by referring to the core portion of known R-gene haplotypes is thus expected since the same R-gene donor is often used for breeding the resistant varieties. Recently, a more robust approach so called targeted amplicon sequencing (TAS) using the next-generation sequencing platform was introduced and optimized for a higher scalability and cost efficient platform for the identification of variants of targeted genes (Meryer et al. 2007; Pertoldi et al. 2009; Bybee et al. 2011). The customized TAS technology provided by companies, e.g., illumina®, enables researchers to efficiently discover and screen genetic variants in hundreds to thousands samples. The future development of diagnostic system using the TAS technology for both R and Avr genes in the rice and rice blast phyto-pathosystem will be promising for implementing a high-throughput analysis applicable for the field monitoring.
The continuous deployment of resistant variety containing same R gene(s) is believed to erode its resistance soon or later, which is speculated to be attributable to the emergence and subsequent dominancy of virulent isolates in the population (Valent and Khang 2010; Wang et al. 2017). In addition to this temporal effect, the spatial effect resulted from the wide distribution of the R genes in different varieties could further increase the risk of defeat of resistance. As reported in this study, Pib and Pi5, which are widely distributed in rice varieties, do not contribute significantly to the varietal resistance any longer. Consistently, the frequencies of their cognate Avr genes in the rice blast population are relatively low, which is most likely attributable to the constant selection pressure posed by the varieties with large planting areas. On the contrary, those R genes, which are yet deployed widely in rice varieties, show relatively strong resistance to the disease, such as Pikm, Pi50, and Pi9. The correlation between the wide distribution of R genes and relatively low resistance frequency conferred by them documented in this study might need more case studies for the further validation. However, the suicide effect of a particular R gene caused by its wide deployment in mega varieties raises a caution for the deployment of R genes. It is worth noting that Pi2 showed a very high frequency of resistance against a rice blast population consisting of 792 isolates, suggesting it was one of promising R genes conferring broad-spectrum resistance against rice blast (Chen et al. 1999). The identification and cloning expedited the use of Pi2 in different rice varieties using MAS (Zhou et al. 2006; Liu et al. 2008; Mi et al. 2018). As of February 2019, Huazhan harboring Pi2 was used as the parental line for a total of 123 hybrid rice varieties, ranking the top restorer line for the breeding of the most number of hybrids (http://www.ricedata.cn/). Due to the highly genetic instability of rice blast pathogen, the efficiency of deployed rice blast R genes in genetically uniform monocultures was speculated to decline continuously over time (Mcdonald and Linde 2002). The finding that the resistance frequency of Pi2 was relatively lower than Piz-t and Pi50 as reported in this study does imply a risk of the breakdown of Pi2 resistance especially along the quick spreading of Huazhan derived rice varieties.
The finding that quite a high frequency of the same non-functional allele at some R-gene loci, such as Pikm1-LTH, Pib-GII, and Pi2/9-V20 respectively at Pik, Pib, and Pi2/9 loci, is surprising and cautious to some degree. It indicated that the genetic diversity of R genes in modern rice varieties is not only limited but also void for resistance. Moreover, it implied that the existing breeding method is cost inefficient and less effective in improving blast resistance since the non-functional haplotypes of the R-gene loci have been unintentionally introgressed and spread in different rice varieties. It is elusive how these non-functional R-gene haplotypes were introduced and retained in the elite rice varieties. However, the selection method for the truly resistant breeding lines including phenotyping by resistance assessment and genotyping using molecular markers could be revisited and refined, which will likely improve the selection efficiency and reliability.
Conclusions
Our study provided an extensive sequence-based diagnosis of R-gene haplotypes in a set of elite rice varieties. The results revealed that quite a number of R-gene haplotypes was non-functional, suggesting that desired R-gene haplotypes were not successfully incorporated in elite rice varieties. Moreover, some R-gene haplotypes were defeated of their resistance due to the low frequency of their cognate Avr genes or haplotypes. Our study envisages an increasing demand on the introgression of effective R genes in rice breeding program through an intensive R gene specific marker selection. We anticipate that the sequence-based diagnosis as described in this study could be widely adopted for validating the presence of rice blast R genes in elite rice varieties and diverse germplasm as well.
Availability of Data and Materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- Avr genes:
-
Avirulence genes
- HMA:
-
Heavy metal-associated
- HR:
-
Highly resistant
- HS:
-
Highly susceptible
- IRBLs:
-
IRRI bred blast resistant lines
- IRRI:
-
International rice research institute
- LRR:
-
Leucine-rich repeat
- LTH:
-
Lijiangxintuanheigu
- MAS:
-
Marker assisted selection
- MR:
-
Moderately resistant
- MS:
-
Moderately susceptible
- NBS:
-
Nuclear binding site
- NCBI:
-
National center for biotechnology information
- NIL-e1:
-
Near isogenic line
- PCR:
-
Polymerase chain reaction
- R genes:
-
Resistance genes
- R:
-
Resistant
- RFs:
-
Resistance frequencies
- RGA:
-
Resistance gene analog
- S:
-
Susceptible
- TAS:
-
Targeted amplicon sequencing
References
Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180:2267–2276
Balachiranjeevi C, Bhaskar N, Abhilash V, Akanksha S, Viraktamath B, Madhav M, Hariprasad A, Laha G, Prasad M, Balachandran S, Neeraja C, Satendra Kumar M, Senguttuvel P, Kemparaju K, Bhadana V, Ram T, Harika G, Mahadeva Swamy H, Hajira S, Yugander A, Pranathi K, Anila M, Rekha G, Kousik M, Dilip Kumar T, Swapnil R, Giri A, Sundaram R (2015) Marker-assisted introgression of bacterial blight and blast resistance into DRR17B, an elite, fine-grain type maintainer line of rice. Mol Breeding 35:151
Bryan GT, Wu K, Farrall L, Jia Y, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2045
Bybee S, Bracken-Grissom H, Haynes B, Hermansen R, Byers R, Clement M, Udall J, Wilcox E, Crandall K (2011) Targeted amplicon sequencing (TAS): a scalable next-gen approach to multilocus, multitaxa phylogenetics. Genome Biol Evol 3:1312–1323
Chaipanya C, Telebanco-Yanoria MJ, Quime B, Longya A, Korinsak S, Korinsak S, Toojinda T, Vanavichit A, Jantasuriyarat C, Zhou B (2017) Dissection of broad-spectrum resistance of the Thai rice variety Jao Hom Nin conferred by two resistance genes against rice blast. Rice 10:18
Chen D, dela Vina M, Inukai T, Mackill D, Ronald P, Nelson R (1999) Molecular mapping of the blast resistance gene, Pi44(t), in a line derived from a durably resistant rice cultivar. Theor Appl Genet 98:1046–1053
Constanzo S, Jia Y (2010) Sequence variation at the rice blast gene Pi-km locus: implications for the development of allele specific markers. Plant Sci 178:523–530
Dodds P, Lawrence G, Catanzariti A, Teh T, Wang C, Ayliffe M, Kobe B, Ellis J (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A 103:8888–8893
Flor H (1971) Current status of gene-for-gene concept. Annu Rev Phytopathol 9:275–296
Fukuoka S, Yamamoto SI, Mizobuchi R, Yamanouchi U, Ono K, Kitazawa N, Yasuda N, Fujita Y, Nguyen T, Koizumi S, Sugimoto K, Matsumoto T, Yano M (2014) Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci Rep 4:1–7
Harlan J (1998) The living fields: our agricultural heritage. Cambridge University Press, Cambridge
Hua L, Wu J, Chen C, Wu W, He X, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, Pan Q (2012) The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet 125:1047–1055
International Rice Research Institute (IRRI) (2002) Standard evaluation system for rice (SES). 5th edn. International Rice Research Institute, Manila, Philippines
Ji Z, Yang S, Zeng Y, Liang Y, Yang C, Qian Q (2016) Pyramiding blast, bacterial blight and brown planthopper resistance genes in rice restorer lines. J Integr Agric 15:1432–1440
Jiang H, Feng Y, Bao L, Li X, Gao G, Zhang Q, Xiao J, Xu C, He Y (2012) Improving blast resistance of Jin23B and its hybrid rice by marker-assisted gene pyramiding. Mol Breeding 30:1679–1688
Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A, Alaux L, Fournier E, Tharreau D, Terauchi R (2012) Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions. Plant J 72:894–907
Khush G, Jena K (2009) Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In: Wang GL (ed) Advances in genetics, genomics and control of rice blast disease. Springer, New York, pp 1–10
Li W, Wang B, Wu J, Lu G, Hu Y, Zhang X, Zhang Z, Zhao Q, Feng Q, Zhang H, Wang Z, Wang G, Han B, Wang Z, Zhou B (2009) The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol Plant-Microbe Interact 22:411–420
Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, Zhou X, Zhu X, Chen Z, Wang J, Zhao W, Ma B, Qin P, Chen W, Wang Y, Liu J, Wang W, Wu X, Li P, Wang J, Zhu L, Li S, Chen X (2017) A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170:114–126
Lin F, Chen S, Que Z, Wang L, Liu X, Pan Q (2007) The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 177:1871–1880
Liu W, Wang F, Jin S, Zhu X, Li J, Liu Z, Liao Y, Zhu M, Huang H, Fu F, Liu Y (2008) Improvement of rice blast resistance in TGMS line by pyramiding of Pi-1 and Pi-2 through molecular marker-assisted selection (in Chinese with English summary). Acta Agron Sinica 34:1128–1136
McDonald B, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Re Phytopathol 40:349–379
Meyer M, Stenzel U, Myles S, Prufer K, Hofreiter M (2007) Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res 35(15):e97
Mi J, Yang D, Chen Y, Jiang J, Mou H, Huang J, Ouyang Y, Mou T (2018) Accelerated molecular breeding of a novel P/TGMS line with broad-spectrum resistance to rice blast and bacterial blight in two-line hybrid rice. Rice 11:11
Ni D, Song F, Ni J, Zhang A, Wang C, Zhao K, Yang Y, Wei P, Yang J, Li L (2015) Marker-assisted selection of two-line hybrid rice for disease resistance to rice blast and bacterial blight. Field Crop Res 184:1–8
Olukayode T, Quime B, Shen Y, Yanoria M, Zhang S, Yang J, Zhu X, Shen W, Tiedemann A, Zhou B (2019) Dynamic insertion of Pot3 in AvrPib prevailing in a field rice blast population in the Philippines lead to the high virulence frequency against the resistance gene Pib in rice. Phytopathology 109(5):870–877
Orbach M, Farrall L, Sweigard J, Chumley F, Valent B (2000) A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12:2019–2032
Pandian B, Joel J, Nachimuthu V, Swaminathan M, Govintharaj P, Tannidi S, Sabariappan (2018) Marker-aided selection and validation of various Pi gene combinations for rice blast resistance in elite rice variety ADT 43. J Genet 97(4):945–952
Pertoldi C, Tokarska M, Wójcik J, Demontis D, Loeschcke V, Gregersen V, Coltman D, Wilson G, Randi E, Hansen M, Bendixen C (2009) Depauperate genetic variability detected in the American and European bison using genomic techniques. Biol Direct 4:48
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang G (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914
Ray S, Singh P, Gupta D, Mahato A, Sarkar C, Rathour R, Singh N, Sharma T (2016) Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front Plant Sci 7:1140
Selisana S, Yanoria M, Quime B, Chaipanya C, Lu G, Opulencia R, Wang G, Mitchell T, Correll J, Talbot N, Leung H, Zhou B (2017) Avirulence (AVR) gene-based diagnosis complements existing pathogen surveillance tools for effective deployment of resistance (R) genes against rice blast disease. Phytopathology 107(6):711–720
Shalini T, Govintharaj P, Ameenal M, Manonmani S, Robin S (2016) Improving blast resistance in parental line of rice hybrid through marker assisted selection. Int J Agric 6:339–346
Skamnioti P, Gurr S (2009) Against the grain: safeguarding rice from rice blast disease. Trends Biotechnol 27:141–150
Su J, Wang W, Han J, Chen S, Wang C, Zeng L, Feng A, Yang J, Zhou B, Zhu X (2015) Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theor Appl Genet 128:2213–2225
Takahashi A, Hayashi N, Miyao A, Hirochika H (2010) Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol 10:175
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822
Valent B, Khang C (2010) Recent advances in rice blast effector research. Curr Opin Plant Biol 13:434–441
Wang B, Ebbole D, Wang Z (2017) The arms race between Magnaporthe oryzae and rice: diversity and interaction of Avr and R genes. J Integr Agric 16:2746–2760
Wang G, Valent B (2017) Durable resistance to rice blast: a resistance gene pair balances immunity against crop yield. Science 355:6328
Wu J, Kou Y, Bao J, Li Y, Tang M, Zhu X, Ponaya A, Xiao G, Li J, Li C, Song MY, Cumagun C, Deng Q, Lu G, Jeon J, Naqvi N, Zhou B (2015) Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9 that triggers Pi9-mediated blast resistance in rice. New Phytol 206:1463–1475
Wu Y, Yu L, Pan C, Dai Z, Li Y, Xiao N, Zhang X, Ji H, Huang N, Zhao B, Zhou C, Liu G, Liu X, Pan X, Liang C, Li A (2016) Development of near-isogenic lines with different alleles of Piz locus and analysis of their breeding effect under Yangdao 6 background. Mol Breeding 36:12
Wu Y, Yu L, Xiao N, Dai Z, Li Y, Pan C, Zhang X, Liu G, Li A (2017) Characterization and evaluation of rice blast resistance of Chinese indica hybrid rice parental lines. Crop J 5:509–517
Xiao G, Borja F, Mauleon R, Padilla J, Yanoria M, Lu G, Dionisui-Sese M, Zhou B (2017) Identification of resistant germplasm containing novel resistance genes at or tightly linked to the Pi2/9 locus conferring broad-spectrum resistance against rice blast. Rice 10:37
Yadav M, Aravindan S, Ngangkham U, Raghu S, Prabhukarthikeyan S, Keerthana U, Marndi B, Adak T, Munda S, Deshmukh R, Pramesh D, Samantaray S, Rath P (2019) Correction: blast resistance in Indian rice landraces: genetic dissection by gene specific markers. PLoS One 14(3):e0213566
Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K, Tosa Y, Chuma I, Takano Y, Win J, Kamoun S, Terauchi R (2009) Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21:1573–1591
Yuan B, Zhai C, Wang W, Zeng X, Xu X, Hu H, Lin F, Wang L, Pan Q (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet 122:1017–1028
Zeng B, Li A, Lyu H (2017) Several features of approval and promotion of Rice varieties in recent years in China (in Chinese with English summary). Hubei Agric Sci 56:21
Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189:321–334
Zhang S, Wang L, Wu W, He L, Yang X, Pan Q (2015) Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci Rep 5:11642
Zhao H, Wang X, Jia Y, Minkenberg B, Wheatley M, Fan J, Jia M, Famoso A, Edwards J, Wamishe Y, Valent B, Wang G, Yang Y (2018) The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun 9:2039
Zhou B, Dolan M, Sakai H, Wang G (2007) The genomic dynamics and evolutionary mechanism of the Pi2/9 locus in rice. Mol Plant-Microbe Interact 20:63–71
Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, Bellizzi M, Wang G (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant-Microbe Interact 19:1216–1228
Acknowledgements
We thank Rice Germplasm Bank of Hunan Academy of Agricultural Sciences for providing part of rice accessions used in this work.
Funding
This work was supported, in part, by grants from the National Natural Science Foundation of China for J.W. (31601384) and G.X. (31801728), and Science and Technology Projects of Guangdong Province for X.Z. (2015B020231003), and National Key Research and Development Program of China for J.W. (2016YFD0101103).
Author information
Authors and Affiliations
Contributions
GX, JW and BZ conceived and designed the experiments. GX, JY, performed the experiments. GX, XZ, JW and BZ analyzed the data. GX, JW and BZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Rice accessions used in this study.
Additional file 2: Table S2.
M. oryzae isolates used in this study.
Additional file 3: Table S3.
Primers used in this study.
Additional file 4: Figure S1.
Validation of R-gene specific primers for PCR amplification.
Additional file 5: Figure S2.
Disproportional distribution of different known R genes in rice varieties.
Additional file 6: Table S4.
Resistance assessment of 54 selected rice varieties against 27 rice blast isolates.
Additional file 7: Figure S3.
Resistance frequency of varieties in 1994 (A) and 2015 (B).
Additional file 8: Figure S4.
Deduction of contribution of known and unknown R genes to the rice varieties.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Xiao, G., Yang, J., Zhu, X. et al. Prevalence of Ineffective Haplotypes at the Rice Blast Resistance (R) Gene Loci in Chinese Elite Hybrid Rice Varieties Revealed by Sequence-Based Molecular Diagnosis. Rice 13, 6 (2020). https://doi.org/10.1186/s12284-020-0367-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-020-0367-x