Abstract
White blood cells, WBCs for short, are an essential component of the human immune system. These cells are our body's first line of defense against infections and diseases caused by bacteria, viruses, and fungi, as well as abnormal and external substances that may enter the bloodstream. A wrong WBC count can signify dangerous viral infections, autoimmune disorders, cancer, sarcoidosis, aplastic anemia, leukemia, tuberculosis, etc. A lot of these diseases and disorders can be extremely painful and often result in death. Leukemia is among the more common types of blood cancer and when left undetected leads to death. An early diagnosis is necessary which is possible by looking at the shapes and determining the numbers of young and immature WBCs to see if they are normal or not. Performing this task manually is a cumbersome, expensive, and time-consuming process for hematologists, and therefore computer-aided systems have been developed to help with this problem. This paper proposes an improved method of classification of WBCs utilizing a combination of preprocessing, convolutional neural networks (CNNs), feature selection algorithms, and classifiers. In preprocessing, contrast-limited adaptive histogram equalization (CLAHE) is applied to the input images. A CNN is designed and trained to be used for feature extraction along with ResNet50 and EfficientNetB0 networks. Ant colony optimization is used to select the best features which are then serially fused and passed onto classifiers such as support vector machine (SVM) and quadratic discriminant analysis (QDA) for classification. The classification accuracy achieved on the Blood Cell Images dataset is 98.44%, which shows the robustness of the proposed work.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
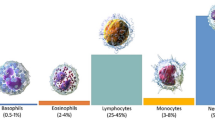
Blood is an integral part of human physiology. It makes up about 7% of an adult’s body weight [1]. It is composed of 55% plasma which allows it to flow freely throughout the body using blood vessels [2]. Centered on the color, size, texture, shape, and composition, the cellular components of blood are separated into three cell types i.e., erythrocytes (red blood cells or RBCs) [3], leukocytes (WBCs) [4, 5], and thrombocytes (platelets) [6]. These elements, when observed under a microscope, have distinct shapes and sizes, with WBC being larger than the others due to the presence of nuclei and cytoplasm in them. This notable feature further divides the WBCs into two types: granulocytes and agranulocytes. Granulocytes are defined by many granules present inside their cytoplasm. These are the most common type of WBC and are produced through granulopoiesis in the bone marrow. There are four sub-types of granulocytes, differentiated based on the color that the granules stain when exposed to a compound dye. These types are neutrophils, eosinophils, basophils, and mast cells. Agranulocytes lack granules in their cytoplasm and are split into two categories—lymphocytes and monocytes [7, 8]. Figure 1 shows one of three WBC types. A sample of 1 µl of human blood contains about 4000–11,000 WBCs. The amount of neutrophils is between 40 and 70%, lymphocytes are between 20 and 45%, monocytes are between 2 and 10%, eosinophils 1 and 6%, and basophils less than 1% [9]. Although they are only 1% of the whole blood volume, their function is very significant and any imbalance can cause serious health complications [10], e.g., leukemia [11], myelodysplastic syndrome, lymphoma, etc. To avoid such complications, diagnosticians and doctors must know the exact count of WBCs in the body. Initial attempts were made by Aldred S. Warthin in 1906 [12], who suggested the use of a diluent that preserved the blood cells. A manual count of the cells was then performed to determine the ratio of cells and their counts. Presently, there are two commonly used ways to determine WBC count—one is to do it manually by using a hemocytometer (microscope staining process) [13] while another involves the use of an automated analyzer.
Complete blood count [14] is a common blood analyzer that provides a count of the unique blood cells present in the body, allowing for the diagnosis of various disorders. For example, a white blood cell count that is lower than normal, called leukopenia [15], can be a sign or cause of marrow cancer, thyroid disorder, an autoimmune disease, typhoid fever, or aplastic anemia, whereas a white blood cell count higher than normal implies that a foreign substance is present in the blood. This condition is called leukocytosis [16] and can cause bone marrow malformation, polycystic ovaries, Addison’s disease [17], and leukemia. To treat WBC disorders, the most important aspect is early diagnosis. For example, early diagnosis of leukemia, which is one of the deadliest forms of cancer, greatly increases the chance of recovery, particularly in children. To achieve this diagnosis, quick and efficient ways are required, so that the quantity and state of the different cells such as monocytes and lymphocytes may be determined, and a conclusion drawn.
The core contributions of the presented research are as under:
-
Channel-wise CLAHE application to improve contrast in the blood cell images.
-
A 45-layer CNN named 4B-AdditionNet, for feature extraction in combination with pre-existing networks.
-
Ant colony optimization (ACO) for feature selection.
-
Linear SVM (LSVM), Cubic SVM (CSVM), quadratic support vector machine (QSVM), linear discriminant analysis (LDA), fine K-nearest neighbor (FKNN), and coarse K-nearest neighbor (CKNN) for classification.
The primary objective of this research is to propose a fully automated, and efficient method for the categorization of WBCs in blood smear images. First, each image of the dataset is preprocessed using CLAHE based image enhancement. This process involves the splitting of the image into three separate images, each representing a single channel from the red–green–blue (RGB) spectrum, trailed by the application of CLAHE on each channel. Second, the feature extraction, using two pre-existing networks in combination with our proposed CNN network named as 4B-AdditionNet is created as a part of this research. Ant colony-based feature selection is utilized to the features extracted by pre-existing networks. Afterward, features fusion is performed to get the benefits of three different types of features. Finally, the classification is performed on different classifiers.
The manuscript is depicted according to the following divisions. An introduction is illustrated in Sect. Introduction. Section Literature Review shows a momentary discussion of current works. The main methodology of this manuscript is encompassed in Sect. Materials and Methods. The experiments and findings are covered in Sect. Results and Discussion. Every experiment contains a unique combination of features from the three networks. Each experiment’s results are analyzed, with relevant information displayed in tables and graphs. Finally, the conclusion and references impart the end of the manuscript.
Literature review
A large amount of work has been done on the classification of blood cells, particularly since the advent of modern CNNs. A steady improvement in accuracy is achieved over the years on most datasets. The importance of this task is evident from the criticality of early detection in the treatment of cancer. Various types of pattern detection and automated computer-based methods have been used in the past, but the speed and accuracy of such methods are low. The feature extraction algorithms such as Speeded Up Robust Features (SURF) [18], Scale Invariant Feature Transform (SIFT) [19], Histogram of Oriented Gradients (HOG) [20], Grey Level Co-occurrence Matrices (GLCM) [21], etc. have been used with moderate success; however, there are still some limitations. Also, CNNs have been used for this purpose and have achieved high accuracy. The process of WBC detection is divided into four main categories—preprocessing, feature extraction, feature selection, and classification.
In preprocessing [22], various techniques have been utilized over time. Prinyakupt et al. [23] performed preprocessing by enhancing the nucleus region of the blood cells by modifying the intensity of different color channels followed by histogram equalization. Bikhet et al. [24] used the technique of median filtering to remove noise from images, followed by thresholding to separate the WBCs from their backgrounds. Karthikeyan et al. [25] used the interpolative Leishman-stained model to remove false areas from blood smear images, followed by re-combining the fragmented parts of the images. Zhong et al. [26] combined the hue–saturation-lightness (HSL) color space with RGB channels for creating a sparse image depiction and later used the sparsity constraint for extracting relevant characteristics from the cell nucleus.
Feature extraction is a process in which important features or attributes of any input data are identified [27,28,29,30,31]. Kutlu et al. [32] combined classes from the Blood Cell Count and Detection (BCCD) and Leukocyte Images for Segmentation and Classification (LISC) datasets to classify five kinds of blood cells by applying Regional CNN (R-CNN) and transfer learning on AlexNet [33], VGG16 [34], GooLeNet [35], and ResNet50 [36]. Toğaçar et al. [37] used the AlexNet model’s FC-8 layer, GoogLeNet model’s loss-3 layer, and ResNet50 model’s FC-1000 layer for feature extraction. These extracted features were then fused in different proportions to achieve a 95.95% accuracy in the classification of WBCs. F.I. Kurniadi et al. [38] used the VGG-16 model in combination with local binary pattern for extracting features. Makem et al. [39] make use of color space transformation using two-color spaces, cyan–magenta–yellow-key (CMYK) and hue-saturation-value (HSV), along with Otsu’s thresholding to segment the blood cell nuclei for feature extraction. Kutlu et al. [32] exploited R-CNN and transfer learning to obtain features using AlexNet, VGG16, GooLeNet, and ResNet50.
Feature selection is the mean through which a large set of extracted features is reduced to a more efficient, smaller set by removing redundant and unproductive features [40]. The accuracy of classification is highly dependent on the feature selection process [41,42,43,44] since the selection of redundant or inefficient features may lead to lower scores and increased computational cost [27]. Gupta et al. [45] improved the accuracy of WBC classification with the optimized binary bat algorithm for dimensionality reduction which resulted in an increase in accuracy compared to [46] and [47]. Sujamol et al. [48] used a genetic algorithm called the inheritable bi-objective combinatorial genetic algorithm (IBCGA) for feature selection in ovarian cancer detection. Ghosh et al. [49] used randomized least absolute shrinkage and selection operator (LASSO) [50] for selecting features in the prediction of cardiovascular disease.
Classification refers to the process during which an input image is assigned a discrete class based on its features [51,52,53,54,55]. This class is the one with the highest probability score amongst all classes. Classification is the final and usually most crucial stage. The success or failure of any algorithm designed for such tasks depends on the classification accuracy. While CNNs can classify an image by themselves since they already have a classification layer, usually using a softmax function [56], there is also the possibility of using a different classifier on extracted features to achieve better results. Baydilli et al. [57] used capsule networks on a small dataset to classify WBCs achieving an accuracy of 96.86%. Banik et al. [58] used nucleus segmentation and a novel CNN to classify the BCCD dataset and accomplished an accuracy of 96%. Gupta et al. [59] used decision-based tree classification on the LISC dataset to achieve a 97.30% accuracy. Almezhghwi et al. [60] used generative adversarial network with deep CNN to classify WBCs to attain an accuracy of 98.8%. A small review of the literature is given in Table 1.
Materials and methods
This unit illustrates a new CNN architecture named 4B-AdditionNet and describes the proposed method along with the steps undertaken to classify WBCs including pretraining of the new network, preprocessing of the dataset using CLAHE, feature extraction using 4B-AdditionNet in combination with ResNet50 and EfficientNetB0, feature selection using ant colony optimization, and classification using multiple classifiers. Figure 2 illustrates an overview of the suggested process. The phases of the projected model are discussed one by one in the upcoming text.
Image enhancement as a preprocessing step
CLAHE [61] is used on the entire dataset to improve the contrast of the dataset images and to make cell bodies more prominent. However, CLAHE can only work on one color channel at a time. For this purpose, this research uses a different technique. First, the image is split into its 3 constituent color channels R, G, and B. Then CLAHE is applied to each of these channels individually, resulting in 3 separate images. These enhanced channel images are then merged back together (see Fig. 3) to produce the final image which has significantly improved contrast than the original.
Proposed CNN-based 4B-AdditionNet
This work contributes a new CNN-based architecture called 4B-AdditionNet (see Fig. 4 for block architecture and Table 2 for structural detail). The backbone structure of this network is like AlexNet; however, a module with four concurrent branches is added after the first convolution layer which aids in improving the accuracy of the network significantly by extracting higher-level features at an earlier stage and feeding it to the lower convolutional layers. The network starts with an input layer that accepts RGB images of size 227 × 227. These images are transferred to the first convolution 2D layer that has 96 filters of size 11 × 11. These filters are applied to the image with a stride of 4 and padding of zero is used. The activation function for this layer is ReLU, which is chased by a cross-channel normalization layer with window size 5. The results are passed through a max-pooling layer with a pool size of 3 × 3 and a stride of 2. The output of this layer is transferred to 4 different groups of layers that work in parallel with each other. This is where the network adapts the Inception block-like approach and becomes wider. This module contains four series of layers working in parallel, with three of them performing different convolutional operations and one layer passing its input along after applying batch normalization. Each convolutional layer in these blocks is tracked by batch normalization and a ReLU layer. The filter sizes of these layers are set to extract the different levels of features simultaneously by utilizing a mix of smaller and larger filters working together, with their results added elementwise in the end by an additional layer. The visualizations of feature maps can be seen in Fig. 5. These results are transferred to a grouped convolution layer. This layer performs multiple convolutions at the same time on the same input. There are two groups of 128 filters of size 5 × 5. The stride value is 1 and padding is 2 on all sides. After passing through the activation function layer, the output is passed to a max-pooling layer with a pool size of 3 × 3 and a stride of 2. The next layer is another convolution layer with 384 filters of size 3 × 3, with a stride of 1 and padding of 1 on each side.
After the activation function, another grouped convolution layer with two groups of 192 filters each, with size 3 × 3, stride 1, and padding 1, work on the input. This is followed by the activation function with feeds into the last grouped convolution layer of this network. This layer has two groups of 128 filters each, with size 3 × 3, stride 1, and padding 1. After activation, a max-pooling layer down-samples the output one last time using a pool size of 3 × 3 with a stride of 2 and padding of 0. At this stage, the convolution process is over, and the data is ready to be flattened and fed to a series of fully connected layers.
The neurons are fully connected layers that come up with full connections to all the activations of the prior layer [62]. The first fully connected layer has an input size of 9216 and an output size of 4096. After the activation function, the data is fed to a dropout layer whose main purpose in a neural network is to prevent overfitting. This dropout layer has a probability of 0.5, which means that there is a 50% chance for every neuron of the previous layer to have its output discarded. The dropout layer is observed by a second fully connected layer with an input and output size of 4096. After the activation function and another dropout layer, the data are fed to the final fully connected layer with an input size of 4096, and outcome size of 100. This output value is reliant on classes of the dataset the network is initially trained on. The final output is forwarded to the softmax layer, which applies the softmax function on the input. The purpose of this function is to transform all input values into a range between 0 and 1. This allows these values to be treated as probabilities since those are always between 0 and 1. The softmax function is given in Eq. 1 below:
where \(z\) is the input vector to the softmax function, \({z}_{i}\) are the elements of the input vector, \({{e}}^{{{z}}_{{i}}}\) is theexponential function applied to each element, \(\sum_{{j}=1}^{{K}}{{e}}^{{{z}}_{{j}}}\) is the normalization, to ensure 0–1 values and \({K}\) is the number of classes.
For this research, 4B-AdditionNet has been learned on the CIFAR-100 [63]. This dataset holds 100 classes, each with 600 images (including train and test) for a total of 60,000 images. The process utilized 50,000 images for training and the remaining 10,000 images for validation purposes.
Feature extraction
In this research, features are extracted using three different CNNs. Features are extracted using the training images from the Blood Cell Images dataset [64] which amount to a total of 9957 images. 4B-AdditionNet extracts 4096 features per image obtained from its FC-2 layer, whereas ResNet50 and EfficientNetB0 extract 1000 features each obtained from the FC1000 and dense|MatMul layer, respectively.
Feature selection
When features are extracted using CNNs, they are often in a large quantity. To reduce the dimensions of those features by selecting a subset, the process of feature selection is used. The feature optimization algorithm chosen for this research is ant colony optimization. It is a probabilistic method for choosing optimal paths. It was first introduced by Dorigo in 1992 [65], based on the seeking comportment of an ant for finding a feasible direction between its associated group and the foodstuff source. In its early days, it was primarily employed to unravel the famous traveling salesman challenge, but after it was applied to various optimization problems as well. The feature selection process followed is completed in the following actions:
-
ACO parameters mentioned in Table 3 are initialized, where
-
a.
the ants are implied by m,
-
b.
the number of iterations is tmax,
-
c.
the evaporation coefficient is ρ with a value of 0 ≤ ρ ≤ 0,
-
d.
the desirability of graph edges is η,
-
e.
α ≥ 0 controls the relative weight of the pheromone,
-
f.
β ≥ 0 controls the weight of η,
-
g.
Q is the amount of initial pheromone concentration.
-
a.
-
For each iteration t, every ant k begins by choosing a random feature. To build a feature subset from the starting feature, the ant follows the probabilistic transition rule given in Eq. 2 below:
$$P_{ij}^k\left(t\right)=\left\{\begin{array}{l}\frac{\tau_{ij}^\alpha\left(t\right).\eta_{ij}^\beta\left(t\right)}{\sum_{l\in S_i^k}\tau_{il}^\alpha.\eta_{il}^\beta\left(t\right)},\forall j\in S_i^k\\0,\,\,{\text{otherwise}}\end{array}\right.,$$(2)where \({S}_{i}^{k}\) represents the feature sets that has not been chosen yet, \({\tau }_{ij}\left(t\right)\) represents the pheromone trail between the features i and j, \({\eta }_{ij}(t)\) represents the heuristic desirability to select feature j while ant k is in the feature i.
-
Assess each contesting feature subset using Eq. 3:
$$\mathrm{Accuracy}=\frac{1}{K} \sum\limits_{i=1}^{K}\frac{1}{2}\left({\mathrm{Accuracy}}_{i}^{\mathrm{Train}}+{\mathrm{Accuracy}}_{i}^{\mathrm{Test}}\right),$$(3)where K denotes the folds set for the K-fold cross-validation procedure to gauge the subset accuracy.
-
The best feature subset is found and the pheromone trail in the feature space is updated.
-
Repeat the process for all ants and finally select the subset with the best accuracy.
Feature fusion
During this process, horizontal concatenation of multiple feature vectors is accomplished to create a single feature vector that can be used for classification. The idea is to merge all features in one feature vector column to possibly help with the reduction of the error rate. In this research, future fusion is used to create multiple feature vectors with different combinations of features from the CNNs to create five experiments. Features from 4B-AdditionNet, ResNet50, and EfficientNetB0 have been fused serially.
Let \({f}_{a}\), \({f}_{r}\), and \({f}_{e}\) denote the three feature vectors acquired from 4B-AdditionNet, ResNet50, and EfficientNetB0. Let \(1\times x\), \(1\times y\), and \(1\times z\) denote the dimensions of \({f}_{a}\), \({f}_{r}\), and \({f}_{e}\) respectively. Then the vectors are defined as:
All obtained feature vectors are fused serially:
Classification
The final step of the process is classification. This process is utilized to predict class labels for the given data. Various classifiers such as SVM [66], LDA [67], and KNN have been used to classify WBCs into four categories. All classifiers utilize fivefold cross-validation. The Linear SVM, Cubic SVM, and Quadratic SVM classifiers utilize a Box Constraint of 1 and a Coding Design of OneVsOne. These predictors are gauged on several performance estimation measures. CSVM is noted with the maximum accuracy while LDA is significantly faster than the competition, with acceptable accuracy.
Results and discussion
The main objective of this study is to utilize a method to classify WBCs with the best possible accuracy. After preprocessing using CLAHE, the proposed network is used along with two other networks to obtain features. Once feature selection is employed to create a feature vector, SVM, LDA, and other predictors are used to evaluate the execution. This portion contains the outcome and assessment of the intended method. First, the details of the execution environment are stated, followed by a brief description of the dataset and the performance evaluation techniques. Finally, the experiments performed are explained in detail.
Execution environment
The training process and experiments were conducted on a Windows-based desktop PC with a 6-core Ryzen 3600 processor from AMD, 16GBs of DDR4-3200 RAM, and a CUDA [68] enabled Nvidia GeForce GTX 1080Ti GPU with 11 GBs of Video RAM (VRAM). The network design, training, and experimentation process were all performed using MATLAB R2020b.
Dataset
The dataset chosen for this study is the Blood Cell Images dataset [69]. It is an augmented version of the BCCD dataset [70] which is publicly available with blood smears divided into 3 classes, RBC, WBC, and Platelets. The Blood Cell Images dataset is an augmented version of the BCCD dataset and contains 12,500 augmented images of blood smears divided into 4 categories—Eosinophil, Lymphocyte, Monocyte, and Neutrophil as shown in Fig. 6. The image size of the augmented set is 320 × 240, bit depth is 24, the number of channels is 3 (R, G, B), and horizontal and vertical resolution depth is 96 dots per inch (dpi). Table 4 shows the details of the dataset samples.
Performance evaluation methods
Evaluating a classification system’s efficiency is a major part of building an accurate classifier. This study implements the most commonly used evaluation metrics such as accuracy (Ac), sensitivity (Se), specificity (Sp), F1 Score, and precision (Pr) [71]. The mathematical formulas for these methods are given in Table 5.
Overview of performed experiments
Using different combinations of features from 4B-AdditionNet, ResNet50, and EfficientNetB0, five tests are performed to determine the top combination of features. The details of these experiments are presented in Fig. 7. fivefold cross-validation is opted in all experiments.
The best results are achieved in the last experiment when using 100 features from 4B-AdditionNet, 400 from ResNet50, and all 1000 from EfficientNetB0.
Test No. 1 (2100 features: 800 4B-AdditionNet + 700 ResNet50 + 600 EfficientNetB0)
This test contains a total of 2100 features with 800 from 4B-AdditionNet, 700 from ResNet50, and 600 from EfficientNetB0. The final feature vector after fusion is of size 9957 × 2100. The Cubic SVM classifier achieved an Ac of 97.58% with a Se of 96.56%, Sp of 98.24%, Pr of 94.85%, and an F1 score of 95.69%, with a runtime of 174.73 s. Table 6 shows the detailed results for all six classifiers. Figure 8 illustrates the training time and Ac for all six classifiers.
Figure 8 illustrates the relationship between training time and Ac when evaluating a 9957 × 2100 feature vector. While Cubic SVM is the most accurate of the six classifiers, LDA strikes the best balance between Ac and speed by being the fastest performing algorithm while having the third-best Ac.
Test No. 2 (1600 features: 700 4B-AdditionNet + 500 ResNet50 + 400 EfficientNetB0)
This test contains a total of 1600 features with 700 from 4B-AdditionNet, 500 from ResNet50, and 400 from EfficientNetB0. The final feature vector after fusion is of size 9957 × 1600. The Cubic SVM classifier achieved an Ac of 97.53% with an Se of 96.60%, Sp of 98.16%, Pr of 94.63%, and an F1 score of 95.60%, with a runtime of 112.71 s. Table 7 shows the detailed results for all six classifiers. Figure 9 illustrates the relationship between training time and Ac for all six classifiers.
Figure 9 depicts the relationship between training time and Ac when evaluating a 9957 × 1600 feature vector. It can be observed again that LDA is the fastest classifier with the third-best Ac, and CSVM remains the most accurate classifier.
Test No. 3 (1050 features: 500 4B-AdditionNet + 250 ResNet50 + 300 EfficientNetB0)
This test contains a total of 1050 features with 500 from 4B-AdditionNet, 250 from ResNet50, and 300 from EfficientNetB0. The final feature vector after fusion is of size 9957 × 1050. The Cubic SVM classifier achieved an Ac of 97.47% with a Se of 96.16%, Sp of 98.34%, the Pr of 95.09%, and an F1 score of 95.62%, with a runtime of 71.86 s. Table 8 shows the detailed results for all six classifiers. Figure 10 illustrates the relationship between training time and Ac for all six classifiers.
Figure 10 shows once again that Cubic SVM remains the most accurate classifier for this dataset, while LDA remains the fastest.
Test No. 4 (650 features: 300 4B-AdditionNet + 150 ResNet50 + 200 EfficientNetB0)
This test contains a total of 650 features with 300 from 4B-AdditionNet, 150 from ResNet50, and 200 from EfficientNetB0. The final feature vector after fusion is of size 9957 × 650. The Cubic SVM classifier achieved an Ac of 96.73% with a Se of 95.31%, Sp of 97.73%, the Pr of 93.37%, and an F1 score of 94.33%, with a runtime of 42.16 s. Table 9 shows the detailed results for all six classifiers. Figure 11 shows the relationship between training time and Ac for all six classifiers.
Figure 11 highlights the sheer speed of LDA, which manages a runtime of only 2.3 s with a respectable Ac of 88.21%. While Cubic SVM takes significantly more time, it is also much more accurate achieving an Ac of 96.73%.
Test No. 5 (1500 features: 100 4B-AdditionNet + 400 ResNet50 + 1000 EfficientNetB0)
This test contains a total of 1500 features with 100 from 4B-AdditionNet, 400 from ResNet50, and 1000 from EfficientNetB0. The final feature vector after fusion is of size 9957 × 1500. The Cubic SVM classifier achieved an Ac of 98.44% with a Se of 97.80%, Sp of 98.87%, the Pr of 96.67%, and an F1 score of 97.23%, with a runtime of 97.29 s. Table 10 shows the detailed results for all six classifiers. Figure 12 illustrates the relationship between training time and Ac for all six classifiers.
In this test, the LDA classifier once again was exceptionally fast and took only 10.24 s to achieve an Ac of 94.96%. It is interesting to note here that LDA remains the fastest performing classifier throughout all five experiments, whereas Cubic SVM remained the most accurate. The confusion matrix for test no. 5 is shown in Fig. 13. There are a total of 2445 correct classifications for eosinophil, 2479 for lymphocyte, 2473 for monocyte, and 2401 for neutrophil.
It can be seen from the confusion matrix given that the two classes most often misclassified are neutrophil and eosinophil. There is a total of 87 cases where the classifier misclassified a neutrophil as an eosinophil, and eosinophil was misclassified as neutrophil. The results for other classes are significantly more accurate. The most accurately classified class is lymphocyte which is incorrectly predicted only four times.
Difference with existing state of the art
In this section, the findings through our experiments are compared to three recent studies on WBC classification. Table 11 contains the details of the methods along with our proposed method.
Discussion
The primary focus of this study is an accurate classification of WBCs. The proposed CNN 4B-AdditionNet is created after extensive testing and experimentation. This network, in combination with ResNet50 and EfficientNetB0, is used to extract features from the Blood Cell Images dataset, which is preprocessed using CLAHE. ACO-based feature selection gives five different combinations of features which are fed to 6 classifiers to determine the accomplishment of the intended method. The findings of five tests shared in Tables 6, 7, 8, 9, 10 portray the accomplishment of the suggested technique. The performance in test no. 5 using 1500 features is found to be the best. It is deduced that while there is a very gradual decline in Ac as the number of features is decreased, however, the relationship is not linear since the best Ac is obtained in test no. 5 using 1500 features, which is better than the results from the test no. 1 with 2000 features. It all comes down to the feature selection process and the number of features used from the different networks. The other interesting observation is that while Cubic SVM’s Ac wavers slightly in all the tests (96.73–98.44), some classifiers have a much higher difference in accuracies across the experiments, e.g., LDA (88.21 – 95.18). Also, while LDA achieves its highest Ac in test no. 1, Cubic SVM does so in test no. 5. Overall, Cubic SVM is the most accurate classifier but also the slowest among the 6, whereas LDA is the fastest and has respectable Ac.
Conclusion
The first conclusion is regarding CNN design that adding width to a network rather than expanding it vertically, leads to far more efficient-performing networks. While this had already been put into effect by other researchers, this study shows that even older networks like AlexNet can be significantly improved just by the introduction of some width-based convolution blocks. It is also construed that top classification accuracies can be obtained on WBC datasets using CNNs as feature extractors without any segmentation done beforehand. In most research work, great emphasis is put on segmentation methods which is a time-consuming task since it requires a lot of fine-tuning to be applicable on entire datasets. This study shows that in the case of WBCs, segmentation can be skipped in favor of simpler preprocessing techniques like CLAHE. Feature fusion technique allows an increase in Ac by using the feature extraction process of multiple networks, and feature selection techniques like ACO enable researchers to tackle the dimensionality curse and keep the feature vectors relatively small even after fusion of multiple feature sets. It can also be deduced that the Blood Cell Images dataset has a slight problem of interclass similarity when it comes to the eosinophil and neutrophil classes which leads to the decrease in the overall Ac of classification. The eosinophil and neutrophil classes have classification accuracies of 97.92% and 96.08% respectively, which is significantly lower than those of lymphocytes and monocytes which are 99.84% and 99.80%, respectively.
Future work
While currently, existing methods have achieved very high Ac in blood cell classification, there is still room for improvement, as shown by this study. Further advances can be made by fusion of the Blood Cell Images dataset with the LISC dataset to add a fifth type of cell to the data, which will create an even more versatile classifier that can perform better across different datasets. Fusion of different networks can also be used to improve performance further, particularly the runtime of classification functions which can be significantly reduced by lowering the number of features.
References
How Much Blood Is in the Human Body? https://www.healthline.com/health/how-much-blood-in-human-body. Accessed: Mar. 05, 2021. [Online]
Loey M, Naman MR, Zayed HH (2020) A survey on blood image diseases detection using deep learning, vol. 11. IGI Global. https://doi.org/10.4018/IJSSMET.2020070102.
Jamil MMA, Oussama L, Hafizah WM, Wahab MHA, Johan MF (2019) Computational automated system for red blood cell detection and segmentation. In: Intelligent Data Analysis for Biomedical Applications: Challenges and Solutions, Elsevier, pp. 173–189. https://doi.org/10.1016/B978-0-12-815553-0.00008-2.
Di Ruberto C, Loddo A, Putzu L (2020) Detection of red and white blood cells from microscopic blood images using a region proposal approach. Comput Biol Med 116:103530. https://doi.org/10.1016/j.compbiomed.2019.103530
Naz I, Muhammad N, Yasmin M, Sharif M, Shah JH, Fernandes SL (2019) Robust discrimination of leukocytes protuberant types for early diagnosis of leukemia. J Mech Med Biol 19(06):1950055
Mahanta LB, Bora K, Kalita SJ, Yogi P (2019) Automated counting of platelets and white blood cells from blood smear images. In: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), vol 11942 LNCS, pp. 13–20. https://doi.org/10.1007/978-3-030-34872-4_2.
Glenn A, Armstrong CE (2019) Physiology of red and white blood cells, vol 20. Elsevier Ltd. https://doi.org/10.1016/j.mpaic.2019.01.001
Shahin AI, Guo Y, Amin KM, Sharawi AA (2019) White blood cells identification system based on convolutional deep neural learning networks. Comput Methods Prog Biomed 168:69–80. https://doi.org/10.1016/j.cmpb.2017.11.015
Pierre RV (2002) Peripheral blood film review: The demise of the eyecount leukocyte differential, vol. 22. W.B. Saunders. https://doi.org/10.1016/S0272-2712(03)00075-1.
Sharif M, Ansari GJ, Yasmin M, Fernandes SL (2018) Reviews of the Implications of VR/AR Health Care Applications in Terms of Organizational and Societal Change. Emerg Technol Health Med Virtual Real Augment Real Artif Intell Internet Things Robot Ind 40:1–19
Jha KK, Dutta HS (2019) Mutual information based hybrid model and deep learning for acute lymphocytic leukemia detection in single cell blood smear images. Comput Methods Programs Biomed 179:104987. https://doi.org/10.1016/j.cmpb.2019.104987
Warthin AS (1906) Experimental ligation of splenic and portal veins, with the aim of producing a form of splenic anemia. Proc Soc Exp Biol Med 4(1):127–128. https://doi.org/10.3181/00379727-4-89
Ozkan A, Isgor SB, Sengul G (2016) Hemositometre Görüntüleri Üzerinde Mikroskop Objektif Ayrimi için Yöntem Önerisi. In: 2016 24th Signal Processing and Communication Application Conference, SIU 2016—Proceedings. pp. 1305–1308. https://doi.org/10.1109/SIU.2016.7495987.
Agarwal R, Sarkar A, Bhowmik A, Mukherjee D, Chakraborty S (2020) A portable spinning disc for complete blood count (CBC). Biosens Bioelectron 150:111935. https://doi.org/10.1016/j.bios.2019.111935
Boxer L, Dale DC (2002) Neutropenia: causes and consequences. Semin Hematol 39(2):75–81. https://doi.org/10.1053/shem.2002.31911
Widick P, Winer ES (2016) Leukocytosis and Leukemia, vol. 43. W.B. Saunders. https://doi.org/10.1016/j.pop.2016.07.007.
Hellesen A, Bratland E, Husebye ES (2018) Autoimmune Addison’s disease—an update on pathogenesis. Ann Endocrinol 79(3):157–163. https://doi.org/10.1016/j.ando.2018.03.008
Bay H, Tuytelaars T, Van Gool L (2006) SURF: speeded up robust features. In: Computer Vision—ECCV 2006, Berlin, Heidelberg, pp. 404–417. https://doi.org/10.1007/11744023_32
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. Int J Comput Vis 60(2):91–110. https://doi.org/10.1023/B:VISI.0000029664.99615.94
Kobayashi T, Hidaka A, Kurita T (2007) Selection of histograms of oriented gradients features for pedestrian detection. pp. 598–607. https://doi.org/10.1007/978-3-540-69162-4_62.
Haralick R, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC-3: 610–621
Amin J, Sharif M, Yasmin M, Fernandes SL (2020) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recognit Lett 139:118–127
Prinyakupt J, Pluempitiwiriyawej C (2015) Segmentation of white blood cells and comparison of cell morphology by linear and naïve Bayes classifiers. Biomed Eng Online 14(1):63. https://doi.org/10.1186/s12938-015-0037-1
Bikhet SF, Darwish AM, Tolba HA, Shaheen SI (2000) Segmentation and classification of white blood cells. ICASSP IEEE Int Conf Acoust Speech Signal Process 4:2259–2261. https://doi.org/10.1109/ICASSP.2000.859289
Karthikeyan MP, Venkatesan R (2020) Interpolative Leishman-Stained transformation invariant deep pattern classification for white blood cells. Soft Comput 24(16):12215–12225. https://doi.org/10.1007/s00500-019-04662-4
Zhong Z, Wang T, Zeng K, Zhou X, Li Z (2019) White blood cell segmentation via sparsity and geometry constraints. IEEE Access 7:167593–167604. https://doi.org/10.1109/ACCESS.2019.2954457
Salau AO, Jain S (2019) Feature extraction: a survey of the types, techniques, applications. In: 2019 International Conference on Signal Processing and Communication (ICSC), NOIDA, India, pp. 158–164. https://doi.org/10.1109/ICSC45622.2019.8938371.
Ansari GJ, Shah JH, Yasmin M, Sharif M, Fernandes SL (2018) A novel machine learning approach for scene text extraction. Future Gener Comput Syst 87:328–340
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: deep convolutional neural networks. Future Gener Comput Syst 87:290–297
Naqi SM, Sharif M, Yasmin M, Fernandes SL (2018) Lung nodule detection using polygon approximation and hybrid features from CT images. Curr Med Imaging 14(1):108–117
Shah JH, Chen Z, Sharif M, Yasmin M, Fernandes SL (2017) A novel biomechanics-based approach for person re-identification by generating dense color sift salience features. J Mech Med Biol 17(07):1740011
Kutlu H, Avci E, Özyurt F (2020) White blood cells detection and classification based on regional convolutional neural networks. Med Hypotheses 135:109472. https://doi.org/10.1016/j.mehy.2019.109472
Krizhevsky A, Sutskever I, Hinton GE (2019) ImageNet classification with deep convolutional neural networks. Available at http://code.google.com/p/cuda-convnet/. [Online]
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. Available at http://www.robots.ox.ac.uk/ [Online]
Szegedy C, et al. (2015) Going deeper with convolutions. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, vol. 07–12-June-2015, pp. 1–9. https://doi.org/10.1109/CVPR.2015.7298594.
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, vol. 2016, pp. 770–778. https://doi.org/10.1109/CVPR.2016.90.
Toğaçar M, Ergen B, Cömert Z (2020) Classification of white blood cells using deep features obtained from convolutional neural network models based on the combination of feature selection methods. Appl Soft Comput J 97:106810. https://doi.org/10.1016/j.asoc.2020.106810
Kurniadi FI, Putri VK (2019) A comparison of human crafted features and machine crafted features on white blood cells classification. J Phys Conf Ser. https://doi.org/10.1088/1742-6596/1201/1/012045
Makem M, Tiedeu A (2020) An efficient algorithm for detection of white blood cell nuclei using adaptive three stage PCA-based fusion. Inform Med Unlock 20:100416. https://doi.org/10.1016/j.imu.2020.100416
Blum AL, Langley P (1997) Selection of relevant features and examples in machine learning. Artif Intell 97(1):245–271. https://doi.org/10.1016/S0004-3702(97)00063-5
Sharif M, Khan MA, Faisal M, Yasmin M, Fernandes SL (2020) A framework for offline signature verification system: best features selection approach. Pattern Recognit Lett 139:50–59
Saba T, Rehman A, Jamail NSM, Marie-Sainte SL, Raza M, Sharif M (2021) Categorizing the students’ activities for automated exam proctoring using proposed deep L2-GraftNet CNN network and ASO based feature selection approach. IEEE Access 9:47639–47656
Saba T, Rehman A, Latif R, Fati SM, Raza M, Sharif M (2021) Suspicious activity recognition using proposed deep L4-branched-ActionNet with entropy coded ant colony system optimization. IEEE Access. https://doi.org/10.1109/ACCESS.2021.3091081
Sharif MI, Khan MA, Alhussein M, Aurangzeb K, Raza M (2021) A decision support system for multimodal brain tumor classification using deep learning. Syst Complex Intell. https://doi.org/10.1007/s40747-021-00321-0
Gupta D, Arora J, Agrawal U, Khanna A, de Albuquerque VHC (2019) Optimized binary bat algorithm for classification of white blood cells. Meas J Int Meas Confed 143:180–190. https://doi.org/10.1016/j.measurement.2019.01.002
Tareef A, Song Y, Cai W, Wang Y, Feng DD, Chen M (2016) Automatic nuclei and cytoplasm segmentation of leukocytes with color and texture-based image enhancement. In: Proceedings—International Symposium on Biomedical Imaging. vol. 2016, pp. 935–938. https://doi.org/10.1109/ISBI.2016.7493418.
Su MC, Cheng CY, Wang PC (2014) A neural-network-based approach to white blood cell classification. World J Sci. https://doi.org/10.1155/2014/796371
Sujamol S, Vimina ER, Krishnakumar U (2021) Improving recurrence prediction accuracy of ovarian cancer using multi-phase feature selection methodology. Appl Artif Intell 35(3):206–226. https://doi.org/10.1080/08839514.2020.1854988
Ghosh P, et al (2021) Efficient prediction of cardiovascular disease using machine learning algorithms with relief and LASSO feature selection techniques. IEEE Access 9:19304–19326. https://doi.org/10.1109/ACCESS.2021.3053759
Zhou C, Wieser A (2018) Jaccard analysis and LASSO-based feature selection for location fingerprinting with limited computational complexity, vol 2018. Progress in Location Based Services, Cham, pp 71–87. https://doi.org/10.1007/978-3-319-71470-7_4
Shah JH, Sharif M, Yasmin M, Fernandes SL (2020) Facial expressions classification and false label reduction using LDA and threefold SVM. Pattern Recognit Lett 139:166–173
Amin J, Sharif M, Yasmin M, Saba T, Anjum MA, Fernandes SL (2019) A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning. J Med Syst 43(11):1–16
Sharif M, Raza M, Shah JH, Yasmin M, Fernandes SL (2019) An overview of biometrics methods. Multimed Inf Secur Tech Appl Handb. https://doi.org/10.1007/978-3-030-15887-3_2
Amin J, Sharif M, Yasmin M, Ali H, Fernandes SL (2017) A method for the detection and classification of diabetic retinopathy using structural predictors of bright lesions. J Comput Sci 19:153–164
Naz J, Sharif M, Yasmin M, Raza M, Khan MA (2021) Detection and classification of gastrointestinal diseases using machine learning. Curr Med Imaging 17(4):479–490
Bridle JS (1990) Probabilistic interpretation of feedforward classification network outputs, with relationships to statistical pattern recognition. In: Neurocomputing. Springer, Berlin Heidelberg, pp. 227–236. https://doi.org/10.1007/978-3-642-76153-9_28.
Baydilli YY, Atila Ü (2020) Classification of white blood cells using capsule networks. Comput Med Imaging Graph 80:101699. https://doi.org/10.1016/j.compmedimag.2020.101699
Banik PP, Saha R, Kim KD (2020) An automatic nucleus segmentation and cnn model based classification method of white blood cell. Expert Syst Appl 149:113211. https://doi.org/10.1016/j.eswa.2020.113211
Gupta D, Agrawal U, Arora J, Khanna A (2020) Bat-inspired algorithm for feature selection and white blood cell classification. In: Nature-Inspired Computation and Swarm Intelligence. Elsevier, 179–197. https://doi.org/10.1016/b978-0-12-819714-1.00022-1.
Almezhghwi K, Serte S (2020) Improved classification of white blood cells with the generative adversarial network and deep convolutional neural network. Comput Intell Neurosci. https://doi.org/10.1155/2020/6490479
Ketcham DJ, Lowe RW, Weber JW (1974) Image enhancement techniques for cockpit displays. US Dept Navy. https://doi.org/10.21236/ADA014928
CS231n Convolutional Neural Networks for Visual Recognition. https://cs231n.github.io/convolutional-networks/#fc (Accessed May 27, 2021).
CIFAR-10 and CIFAR-100 datasets. Available at https://www.cs.toronto.edu/~kriz/cifar.html. Accessed: Mar. 09, 2021. [Online].
Blood Cell Images|Kaggle.. Available at. https://www.kaggle.com/paultimothymooney/blood-cells. Accessed: Mar. 10, 2021. [Online]
Dorigo M (1992) Optimization, learning and natural algorithms. PhD Thesis Politec. Milano. Available: https://ci.nii.ac.jp/naid/10000136323/en/Accessed: May 28, 2021. [Online]
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20(3):273–297. https://doi.org/10.1007/BF00994018
Fisher RA (1936) The use of multiple measurements in taxonomic problems. Ann Eugen 7(2):179–188. https://doi.org/10.1111/j.1469-1809.1936.tb02137.x
Ghorpade J, Parande J, Kulkarni M, Bawaskar A (2012) GPGPU processing in CUDA architecture. Adv Comput Int J 3(1):105–120. https://doi.org/10.5121/acij.2012.3109
Blood Cell Images. https://kaggle.com/paultimothymooney/blood-cells (Accessed Apr. 14, 2021).
Shenggan/BCCD_Dataset: BCCD (Blood Cell Count and Detection) Dataset is a small-scale dataset for blood cells detection. Accessed: Mar. 06, 2021. [Online]. Available at https://github.com/Shenggan/BCCD_Dataset
Junker M, Hoch R, Dengel A (1999) On the evaluation of document analysis components by recall, precision, and accuracy. In: Proceedings of the International Conference on Document Analysis and Recognition, ICDAR, pp. 717–720. https://doi.org/10.1109/ICDAR.1999.791887.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shahzad, A., Raza, M., Shah, J.H. et al. Categorizing white blood cells by utilizing deep features of proposed 4B-AdditionNet-based CNN network with ant colony optimization. Complex Intell. Syst. 8, 3143–3159 (2022). https://doi.org/10.1007/s40747-021-00564-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00564-x