Abstract
Multiclass classification of brain tumors is an important area of research in the field of medical imaging. Since accuracy is crucial in the classification, a number of techniques are introduced by computer vision researchers; however, they still face the issue of low accuracy. In this article, a new automated deep learning method is proposed for the classification of multiclass brain tumors. To realize the proposed method, the Densenet201 Pre-Trained Deep Learning Model is fine-tuned and later trained using a deep transfer of imbalanced data learning. The features of the trained model are extracted from the average pool layer, which represents the very deep information of each type of tumor. However, the characteristics of this layer are not sufficient for a precise classification; therefore, two techniques for the selection of features are proposed. The first technique is Entropy–Kurtosis-based High Feature Values (EKbHFV) and the second technique is a modified genetic algorithm (MGA) based on metaheuristics. The selected features of the GA are further refined by the proposed new threshold function. Finally, both EKbHFV and MGA-based features are fused using a non-redundant serial-based approach and classified using a multiclass SVM cubic classifier. For the experimental process, two datasets, including BRATS2018 and BRATS2019, are used without increase and have achieved an accuracy of more than 95%. The precise comparison of the proposed method with other neural nets shows the significance of this work.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brain tumor is one of the most terrifying diseases in the present era [1, 2]. The collective behavior of the abnormal cells in the brain is a one of the common reasons for its occurrence. [3]. Two stages of brain tumors including primary and secondary are reported in the relevant literature [4]. In the primary stage, the tumor size is small, and in the biological term, it is called benign. In the secondary stage, tumors spread from other parts of the body and its size is larger than benign, and it is called malignant [5]. According to the National Brain Tumor Society, in USA, approximately 700,000 patients are suffered from brain tumor disease. Of those, 69.8% are benign, whereas rests of 30.2% are malignant in nature. According to the report, the survival rate of the patients is 36% only. In 2020, approximately 87,000 patients are diagnosed with brain tumor [6]. In 2021, an estimated number of diagnosed brain tumor patients are 84, 170. The number of diagnosed adults above age 40 will be 69, 950. Based on high mortality rate of brain tumor, it is divided into two stages—HGG (high-grade glioma) and LGG (low-grade glioma). Moreover, the LGG survival rate is fast as compared to HGG. The survival rate of HGG is approximately 2 years; therefore, it is required a fast treatment [7].
For the treatment of brain tumors, different techniques are used in the clinics [8]. In the benign stage, radiotherapy is useful and a patient can survive without any surgery [9]. On the other side, the cancerous stage is harmful and can be treated through chemotherapy and radiotherapy [10]. Hence, benign tumors are typically slow in spreading as compared to malignant tumors. However, in either case, diagnosis is crucial and it needs expert radiologists [11]. The more recent imaging technology shows a huge success in the area of medical imaging for the diagnosis and detection of dangerous human diseases such as brain tumors [12], skin cancer [13], stomach cancer [14, 15], lung cancer [16], blood cancer [17], and name a few more [18,19,20,21]. For brain tumors, MRI (Magnetic Resonance Imaging) and CT (Computed Tomography) scans are more useful imaging technologies [22]. However, MRI scans are more useful based on tumor texture and shape information as compared to CT images [23]. Through MRI, the size, shape, and location of the detective tissues can be easily computed. These techniques also have few flaws such as huge computational time and cost [24, 25].
For early brain tumor detection and classification, a computer-aided diagnosis (CAD) system may helpful for the second opinion of the radiologists in the clinics [26, 27]. A simple CAD system consists of few important steps in a sequential manner such as preprocessing of original MRI scans, feature extraction of preprocessed MRI scans, feature reduction, and finally classification using a supervised learning algorithm [28]. In the preprocessing step, the original MRI scans are improved where tasks like contrast enhancement and noise removal are performed. This is important for manual/classical feature extraction. The classical features are shaped geometric, texture (LBP and GLCM), and point (SIFT). However, these features are not efficient for high-dimensional datasets. A few researchers introduced feature reduction techniques for the fast execution of a CAD system [29]. However, it is not a good way as it may neglect and drop important features. The final features are classified using supervised learning algorithms such as KNN, Naïve Bayes, etc. [30]. This type of CAD system is not supportive of multiclass classification problems. Recently, the entrance of deep learning in the area of medical imaging showed great success for the classification task, especially the multiclass classification problem with improved accuracy. In this regard, the deep learning algorithms are successfully applied for large-dimensional datasets. For brain tumor, the famous datasets are collected from BRATS [31, 32]. This dataset includes four tumor types for each patient, such as T1-weighted tumor, T1 contrast-enhanced tumor, T2-weighted tumor, and Flair, as shown in Fig. 1. In the figure, it is shown that most of the image regions are similar to each other and there is a high chance of misclassification. Moreover, the textural and shape information of these tumors are similar to each other.
In this work, a new automated system using deep learning is proposed, which considers the aforementioned problems of multiclass brain tumor classifications such as similarity among tumor types, reduction of important features, and high-dimensional datasets. In the proposed method, we extract deep learning features without employing the preprocessing step (contrast stretching). Major contributions of this research work are as follows:
-
Fine-tuned a pre-trained deep learning model Densenet201 and trained using a deep transfer learning. The training is conducted on imbalanced data. From the trained model, features are extracted from the average pool layer which represents the highly deep information of each tumor type.
-
Proposed a new feature selection approach named Entropy–Kurtosis-based High Feature Values (EKbHFV). This approach considers number of iterations (total features), and in each iteration, features are validated using Fine-KNN-based fitness function.
-
Modified the Genetic Algorithm (MGA) for the best feature selection using standard deviation. The fitness in each iteration is calculated using Euclidean Distance. If fitness is not meet then performed a cross-over and mutation. In the last, the selected vector is further passed in a threshold function and removes the redundant features.
-
Selected features of both EKbHFV and MGA are fused using a non-redundant serial-based approach. The final features are classified using a multiclass cubic SVM classifier.
The related work of this article is set out in the section “Related work”. The proposed methodology, consisting of a fine-tuned model, extraction features and fusion, is presented in the section “Proposed work”. Results and comparisons with recent techniques are discussed in the section “Experimental results and analysis”. Finally, the conclusion of this work is set out in the section “Conclusion”.
Related work
In the literature, several methods are proposed for brain tumor detection and classification. Most of them focus on binary class classification such as benign and malignant [33, 34]. However, the binary class classification is easy due to the easy interpretation of tumor shape and texture [35, 36]. The multiclass classification problem is difficult due to high similarity among tumor types. Sharif et al. [6] presented the technique to minimize the detection process of the brain tumor and feature selection was the major objective of this research. In the study, a deep learning method is used to recognize the brain tumor. Initially, the contrast enhancement through the saliency method is applied for tumor detection. Later, deep learning features are extracted and optimized using PSO. Two datasets including BRATS2017 and BRATS2018 were used. The accuracies of 83.73% (core tumor), 93.7% (whole tumor), and 79.94% (enhanced tumor) for the BRATS2017 dataset is reported, whereas the accuracy of other dataset BRATS2018 is reported 88.34% (Core tumor), 91.2% (whole tumor), and 81.84% (enhanced tumor). Moreover, the classification process is also applied to other datasets like BRATS2013, BRATS2014, BRATS2017, and BRATS 2018, and reported an average accuracy of 92%. In another research, Narmatha et al. [37] presented segmentation and classification techniques with the help of a fuzzy brain-storm optimization algorithm. In this method, the storm optimization provides the highest priority from the target cluster in the brain. Several iterations of the fuzzy technique help to get the optimal solution. The BRATS2018 dataset was used for the experimental process and an accuracy of 93.85% is reported. Rehman et al. [12] presented a method for the automated detection of brain tumors using a deep learning. The study is useful for the microscopic detection of the tumor. In this method, for the extraction of the brain tumor, the new 3D CNN model is designed. Then, a pre-trained Model of CNN was trained for feature extraction. In the last, optimal features were selected and performed experiments on BRATS 2015, 2017, and 2018. On these three datasets, the accuracies of 98.32%, 96.97%, and 92.67% are reported, respectively. Rehman et al. [38] proposed a framework using three different architectures such as Alexnet, VGGNet, and GoogLeNet. The authors used the transfer learning techniques on each neural net for training. Before training, they performed data augmentation for better classification accuracy. The main purpose of the work is to reduce the problem of over fitting with a large number of data. For the dataset of this performance, MRI slices of fine and freeze were used and gained the best accuracy of 98.69% using VGG architecture. Khan et al. [25] presented the automated model for the classification of brain tumors. This model consists of five major steps. Initially, an edge-based histogram and DCT (discrete cosine transform) transform were used for stretching of linear contrast of MRI scans. In the next step, DL is used with pre-trained models—VGG16 and VGG19 for feature extraction. In the third step, the best features were selected and classified using ELM (Extreme Learning Machine) classifier. This method was performed on the widely known datasets BraTs2015, 2017, and 2018, and accuracy of 97.8%, 96.9%, and 92.5% is reported, respectively. Mzoughi et al. [39] presented the technique for the easiness of the neuroradiology. The main objective of this research was brain tumor detection with volumetric 3D MRI. To make the process more efficient, the authors used Multiscale 3D CNN architecture to classify the tumors. The proposed method has ability to reduce the weight of local and global information via small kernels. Furthermore, the data augmentation approach is employed for better training of the model. In the end, they showed the impact of data augmentation with the help of experimental results.
Proposed work
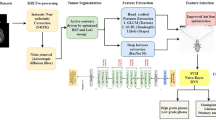
A new efficient deep learning-based framework is presented in this work for multiclass brain tumor classification. In the proposed architecture, the imbalanced data are employed instead of balanced data for training of a fine-tuned Densenet201 deep learning model. Features are extracted from the GAP layer for the classification. However, some of the extracted features of this layer are not useful for accurate classification purposes; therefore, we proposed a new feature selection approach name EKbHFV. In parallel, we modify the GA for the best feature selection. The selected features of both techniques are finally fused using a non-redundancy-based fusion approach. In the end, multiclass CSVM is employed for the final classification. The main architecture of this proposed method is illustrated in Fig. 2.
Convolutional neural network
Convolutional Neural Network (CNN) is one of the famous deep learning architectures, where each layer is connected in a feed-forward way [40]. In CNN architecture, end-to-end learning is performed for the hierarchical representation of an input image. Many layers are added in this architecture for the extraction of local and global information of each image. Recently, these models are more useful for object classification [41], surveillance [42], and medical imaging [43]. A representative CNN architecture consists of several layers, and a few famous layers are: (1) convolutional; (2) ReLu; (3) pooling, and (4) fully connected (FC). CNN model depends on the three main layers that are convolution, pooling, and fully connected layer. Also to overcome the problem of overfitting and generalization, a dropout and batch normalization layers are also added in a CNN architecture [44]. Hence, the abstract-level features are extracted and return output score at the end of this architecture.
A convolutional layer is one of the most important layers in a CNN architecture, which consists of trainable weights. Through these weights, the spatial features (edges and contour) and high-level features (complex structures) are learned. This layer receives an input of dimension \(h\times w\times 3\) and returns the feature maps as output. All feature maps are the dot product of a particular field and weights. These weights are captured the features information of each class during the learning process. The learning process is performed in backpropagation and SDG (gradient descent). Mathematically, this layer is formulated as follows:
where \({\psi }_{k}^{i}\) represent the output of a convolutional layer, \({\beta }_{k}^{i}\) denoted the bias term, \({\mathbb{w}}_{k,j}^{i}\) represent the weight of the convolutional layer, \({\mathbb{x}}_{j}^{i-1}\) represent input terms, and \(\mathrm{Rel}(.)\) denotes non-linearity ReLu activation function. These weights are updated using the output of the previous layer set as an input of the next layer. This function is defined as follows:
This function converts negative weights into zero and considers positive weights as it is. A max-pooling layer is added in a CNN architecture to calculate the maximum values of a given rectangle, where the rectangle is based on the filter size. Each rectangle is moved based on the stride value. This layer is useful to reduce the number of features (weights). The parameters of this layer are the filter dimension, padding mode, and stride. Mathematically it is presented as follows:
The output of these layers in the form of two-dimensional arrays is converted into a one-dimensional array in a fully connected layer. The features of this layer are finally classified using Softmax classifier:
where \({\psi }_{k}^{i}(\mathrm{FC})\) represents the output of the FC layer which is used as an input in Softmax and \(K\) represents the number of classes.
Pre-trained DenseNet 201 model
In traditional CNN, the connectivity of layers was very complex and the data transferred through these layers. In this process, it is a high chance of error occurrence and an increase in the computational cost. In the Resnet architecture, the problem of complexity is resolved and made easier with the skipping layers. The minimum two layers are skipped in this architecture. Densenet improves the model due to the concatenation of the features, in which all features are connected sequentially (linear form). This is a better approach than that summation of feature from the output layer and used as an input for next. The formulation of this process is defined as follows:
where \(m\) stands for the layer index, \(Z\) means the non-linear operation, and \({S}_{m}\) represents the feature of the\( m\mathrm{th}\) layer. Original DenseNet201 architecture [45] is illustrated in Fig. 3. In this figure, it is illustrated that, originally, this model trained on the ImageNet dataset. In each block, the skipping option is available which faster the classification process. Each block consists of many layers such as convolution, ReLu, batch normalization, and pooling. In the end, the features of the transition layer are classified using Softmax classifier.
DenseNet 201 architecture for image classification [45]
In this work, we fine-tuned the DenseNet201 pre-trained model for multiclass brain tumor classification. For this purpose, the last two layers are removed, and add a new FC layer which includes four types of brain tumor. The same weights are used in the fine-tuned model and trained this model using transfer learning. The detail of transfer learning in the next section (Fig. 4).
Transfer learning-based feature extraction
Transfer learning (TL) [46] is one of the best methods to reuse a pre-trained deep learning model for another task. Normally, the TL technique is adopted for training a model for another task using fewer training data; in this approach, training a target model using prior trained knowledge of the related task. This process is helpful to give an accurate solution for fewer training samples. Hence, we can say that TL is useful when target training data are less as compared to source training data.
Consider, the source data with the learning task \({P}_{\begin{array}{c}s \\ \\ \end{array}}= \left\{\left({d}_{1}^{s},{e}_{1}^{s}\right),...,\left({d}_{i}^{s},{e}_{i}^{s}\right),\dots ,\left({d}_{n}^{s},{e}_{n}^{s}\right)\right\}\) and source labels are \({{{P}_{L}=L}_{P},L}_{S},\left({d}_{m}^{s},{e}_{m}^{s}\right)\epsilon R\). Also, the target data with the learning task \({J}_{\begin{array}{c}T \\ \\ \end{array}}= \left\{\left({d}_{1}^{t},{e}_{1}^{t}\right),...,\left({d}_{i}^{t},{e}_{i}^{t}\right),\dots ,\left({d}_{m}^{t},{e}_{m}^{t}\right)\right\}\) and target labels \({{{J}_{L}=L}_{J},L}_{T},\left({d}_{n}^{t},{e}_{n}^{t}\right)\epsilon R\). Where \(n\ll m\) and \({e}_{1}^{P} \,and \, {e}_{1}^{T}\) are the labels of training data. The target goal of TL is to make the more learnable of \({J}_{\begin{array}{c}T \\ \\ \end{array}}\) by the combined knowledge \({P}_{\begin{array}{c}s \\ \\ \end{array}}\) and \({J}_{T}\). Hence, transfer learning can be explained as:
Visually, the process of transfer learning is illustrated in Fig. 5. In this figure, it is illustrated that the source model DenseNet201 is trained on the ImageNet dataset, whereas the number of labels is 1000. By employing transfer learning, knowledge is transfer to a fine-tuned target model. Later, the SGD learning method is used for training this new target model for brain tumor classification. For training, the learning rate is 0.0001, mini-batch size is 64, and the number of epochs is 100. For the target model, two datasets—BRATS2018 and BRATS2019—are utilized. In the end, features are extracted from the global average pool layer for further processing. On this layer, the feature vector (\({\Psi }_{k}^{N}\)) dimension is \(N\times 2048\), where \(N\) represents the number of training samples. As in this work, we consider the 50:50 approach for evaluation of this proposed architecture; hence, \(N\) are 50% and 50%, respectively.
MGA-based feature selection
GA is mostly used in artificial intelligence and machine learning for getting the optimal solution. Many other algorithms are used to generate output, but GA is designed to solve complex problems with larger numbers of population size. There are six steps to solve the GA problem as Initial Population, Calculate fitness, Selection, Crossover, Mutation, and Optimal Solution. In the first, the initial population is initialized. Next, the fitness value is calculated with every individual population. The third step is to select the best fitness values, because it is directly proportional or fitness value. Only the better fitness value generates a better optimal solution, and this is the genetic operation. In the fourth and fifth steps, a new population is generated using the swapping method. Crossover and Mutation are the primary processes in GA to generate the new best fitness values from the parent’s population. If we do not get the highest fitness value, it will be in continuity until the stopping criteria fulfill.
In this article, we modify the output of GA and called modified GA (MGA). In the MGA, the output of GA is passed in a threshold function to remove the redundant features. The advantage of this step is to minimize the computational time and increase accuracy. Let \({\Psi }_{k}^{N}\) is an \(N \times 2048\)-dimensional feature vector, extracted from the fine-tuned DenseNet201 model. Consider, \(\mathrm{M}\) is population size and value of \(M=80\) in this work. The chromosome \(\Psi \) represent the individual features of \({\Psi }_{k}^{N}\). In the initial population, \(P =\left\{{\mathbb{c}}_{1},{\mathbb{c}}_{2},\dots , {\mathbb{c}}_{M}\right\}\), where all the genes of \({\mathbb{c}}_{1}\) are ‘1’. This shows that all descriptors have equal weights. For the other individuals, the random values are generated as \({\phi }_{k}\in [0 1]\) and \(k=1,2,3,\dots\Psi \). In the next step, the fitness value is calculated for each individual using Euclidean Distance (ED). Mathematically, the ED between two individuals is computed as follows:
After this step, the genetic operations are performed for the searches of better solutions (features). The genetic operations are Roulette Wheel-based selection, cross-over, and mutation. The selection operation selects the best individuals based on fitness values. For criteria of new population, the top 70% fitness values are selected; otherwise, generate a new population using crossover and mutation operation.
For a crossover, two individuals are required with a crossover rate \({C}_{r}\). In this work, we utilized a double crossover with the \({C}_{r}=0.5\). Mathematically, the crossover operation is defined as follows:
After this operation, a mutation operation is applied on the crossover individuals with a very small mutation rate \({m}_{r}=0.2\). Four genes of individuals are randomly selected for the mutation operation. Mathematically, the mutation operation is defined as follows:
where \(r\) is a random value between \([0 1]\), \(t\) represents iteration number, and \(\mathrm{It}\) represents the total iterations which are 500 in this work. This process is continued until the max iterations are completed. After the selection of the best features of GA, a new threshold function is proposed. Through this new function, the redundancy among features is removed and decrease the computational cost. Mathematically, this function is formulated as follows:
This formulation shows that of \(i\mathrm{th}\) and \(i\mathrm{th}+1\) features are match each other than select only \(i\mathrm{th}+1\) and ignore the \(i\mathrm{th}\) feature. In this work, we obtained a final selected feature vector of dimension \(N\times 952\) and denoted by \({\mathrm{GA}}_{k}^{N}\).
Entropy–Kurtosis-based feature selection
Consider \({\Psi }_{k}^{N}\) is original deep extracted feature vector of dimension \(N\times K\), and \(K\) represents the extracted features dimension and \(K\in {\mathbb{R}}\). Consider \({\Psi }_{rs}^{N}\) is a resultant selected feature vector of dimension \(N\times rs\). Initially, the Shanon Entropy is computed from each pair of features and obtained an entropy vector of same dimension \(N\times rs\).
Similarly, the Kurtosis value is computed for each pair and gets the peak frequency feature for the selection of entropy features:
Based on the \(\mathrm{Kur}\left(k\right)\), the peak frequency value is obtained and then compared with \(E(K)\) for feature selection at the first phase:
From this function, the entropy features whose values are greater or equal than peak kurtosis value are selected and the rest of them are removed. Then, the selected features are validated through fine-KNN classifier-based fitness function. We added this process in a loop and number of total iterations are 20. After the max iterations, the best accuracy based vector is obtained. In this work, the selected vector length is \(N\times 738\) for iteration 9 and denoted with \({\mathrm{Ent}}_{k}^{N}\), where \({\mathrm{Ent}}_{k}^{N}\in {\Psi }_{E}^{N}\left(k\right)\).
Features fusion and classification
Finally, the selected features of MGA approach and EKbHFV approach are fused in a serial-based method and then remove the redundant information among them. In this technique, initially serially fusion is defined as follows:
Then, the features in this resultant vector are removed by comparing each other and consider only one feature of same values. This comparison process is continued for all features. In the last, we obtained an updated resultant feature vector \({\mathrm{Fus}}_{k}^{N}\) of dimension \(N\times 1310\). The dimension of this vector shows that almost 400 features are redundant which are removed in the comparison phase. As a final, the multiclass cubic SVM is utilized for the final features classification. The results are discussed in the next section.
Experimental results and analysis
Experimental setup
The experimental process of proposed multiclass brain tumor classification method is presented in this section. Two datasets are considered for the evaluation of proposed method- BRATS2018 and BRATS2019. These datasets are more prominent and mostly useful for this domain. The main target in this work is to achieve improved accuracy and minimizing the computational cost. The datasets have two modalities—LGG and HGG—where each modality consists of four stage tumors such as T1-weighted, T1CE, and T2-weighted and Flair (Sample images can be seen in Fig. 6). Several classifiers are used for the analysis of experimental phase. The famous ones are SVM, Fine KNN, and Ensemble Trees. For the evaluation of each classifier performance, various measures are calculated such as recall rate, precision rate, F1-Score, AUC, accuracy, and testing time (sec). This method is implemented on MATLAB2020a using Core i7 Desktop Computer having 16 GB of RAM and 16 GB GPU.
Results and analysis
BRATS2018 Dataset Results: We split dataset into 50/50 ratio, which means that 50% data are used for training and remaining 50% is for testing with 10-Fold cross-validations. Several classifiers are used for the evaluation as listed in Table 1. This table represents the classification results and achieved the best accuracy for Cubic SVM classifier that is 99.9% for HGG and 99.1% for LGG. The other measures of CSVM are recall rate, precision rate, and F1-Score of values 99.9%, 99.8%, and 99.8% for HGG modality. Similarly, these measures are also computed for LGG modality and computed values are 99%, 99%, and 99%, respectively. The noted time of CSVM during the testing process is 151.67 (sec) for HGG and 246.01 (sec) for LGG. The minimum noted time for this experiment is 72.049 (sec) for HGG and 77.288 (sec) for LGG on Linear Discriminant classifier. However, the accuracy of this classifier is 99.7% for HGG and 97.7% for LGG, which is less as compared to CSVM. Moreover, the CSVM performance is also compared with a few well-known classification algorithms in this table and shows that the CSVM results are better.
Table 2 represents the results of BRATS2018 datasets after fusion of optimal selected features. This table shows the best noted accuracy of 99.7% and 98.8% on CSVM for HGG and LGG, respectively. The other noted measures are recall rate, precision rate, and F1-Score of values 99.7%, 99.7%, and 99.7% for HGG and 98.7%, 98.7%, and 98.7%, for LGG, respectively. The noted computational time during the testing process is 88.455 (sec) and 113.40 (sec) which is minimized as compared to the noted time in Table 1. The minimum noted time in Table 2 is 22.946 (sec) and 20.938 (sec) for Linear Discriminant Analysis. Previously (in Table 1), this time was 72 (sec) and 77 (sec). This time shows that the selection of optimal features and fusion these features not only maintain the accuracy, but, on the other side, decrease the testing computational time. The comparison of proposed accuracy on CSVM is also compared with the other classification methods mentioned in Table 2. From results, it can be analyzed that the CSVM performance is overall better. Also, the CSVM recall rate can be validated through confusion matrix illustrated in Figs. 7 and 8. Figure 7 shows the confusion matrix of CSVM for HGG modality, whereas the Fig. 8 shows the confusion matrix of LGG modality.
BRATS2019 dataset results
Similar to BRATS2018, we split dataset into 50/50 ratio, which means that 50% data are used for training and remaining 50% are for testing with tenfold cross-validations. Results are given in Table 3 (without feature fusion). This table represents the best achieved classification accuracy is 99.9% and 99.5% cubic SVM. The 99.9% accuracy is achieved for HGG modality and 99.5% for LGG modality. The other measures of CSVM are recall rate, precision rate, and F1-Score of values 99.9%, 99.9%, and 99.9% for HGG modality. Similarly, these measures are also computed for LGG modality and computed values are 99.5%, 99.5%, and 99.5%, respectively. The noted time of CSVM during the testing process is 144.86 (sec) for HGG and 188.36 (sec) for LGG. The minimum noted time for this experiment is 69.895 (sec) for HGG and 68.577 (sec) for LGG on Linear Discriminant classifier. However, the accuracy of this classifier is 99.7% for HGG and 98.7% for LGG, which is less as compared to CSVM. Moreover, the CSVM performance is also compared with a few well-known classification algorithms in this table and shows that the CSVM results are better.
Table 4 represents the results of BRATS2019 datasets after fusion of optimal selected features. This table shows the best noted accuracy of 99.8% and 99.3% on CSVM for HGG and LGG, respectively. The other noted measures are recall rate, precision rate, and F1-Score of values 99.8%, 99.8%, and 99.8% for HGG and 99.3%, 99.3%, and 99.3%, for LGG, respectively. The noted computational time during the testing process is 80.285 (sec) and 104.56 (sec) which is minimized as compared to the noted time in Table 3. The minimum noted time in Table 4 is 23.231 (sec) and 20.631 (sec) for Linear Discriminant Analysis. Previously (in Table 3), this time was 69 (sec) and 68 (sec). This time shows that the selection of optimal features and fusion these features minimize the testing computational time and also increase the classification accuracy. The comparison of proposed accuracy on CSVM is also compared with other classification methods mentioned in Table 4. From results, it can be analyzed that the CSVM performance is overall better. Also, the CSVM recall rate can be validated through confusion matrix illustrated in Figs. 9 and 10. Figure 9 shows the confusion matrix of CSVM for HGG modality, whereas the Fig. 10 shows the confusion matrix of LGG modality.
Comparison
In this section, we compare the propose method accuracy with other neural nets and also analyze the overall system performance. Before the selection, the maximum accuracy was 93% for BRATS2018 and 92% for BRATS2019. Moreover, the testing computational time was more than 500 (sec). Tables 1 and 3 show the classification results without using features fusion. From these tables, it is show that the accuracy and time efficiency of the proposed method is improved after employing feature selection techniques. Furthermore, it is improved after the fusion results, as presented in Tables 2 and 4. The comparison of each dataset is also conducted in Tables 5 and 6. In Table 5, the achieved accuracy for VGG, AlexNet, ResNet101, and Inception V3 for HGG is 95.3%, 93.6%, 96.9%, and 97.4%. Similarly, for LGG, the achieved accuracy was 92.7%, 91.8%, 95.2%, and 96.9%. For the proposed method, achieved accuracy is 99.7% and 98.8 using BRATS2018 dataset which shows the improvement of this approach. Likewise, Table 6 shows the comparison of BRATS2019 dataset and described that the performance of proposed method is improved.
Conclusion
This work presents a deep learning automated system for the classification of brain tumors into four types such as T1W, T1CE, T2W, and Flair. Brain MRI scans are more useful imaging technology for the analysis of brain tumors; therefore, this system may be useful for the second opinion of radiologists. Multiclass classification of brain tumors is a complex and difficult task due to the high similarity between the tumor stages. Also, existing systems work well for balancing datasets, which is not a good way, because several images are ignored during the learning process. The main strength of this proposed method is the selection of the most optimal features using MGA and Entropy–Kurtosis-based techniques. These proposed techniques improve the accuracy of the system and also reduce the time of classification. The second strength of this work is the fusion of the optimal features to further improve the proposed accuracy. The experimental process shows that the proposed method shows a significant improvement in the datasets selected. The experimental process for BRATS2019 datasets with more recent deep learning methods will be conducted in the future. The main limitation of this work is the reduction of certain important features which have an impact on the accuracy of the system. In addition, the fusion process increases the computational time.
References
Al-Okaili RN, Krejza J, Wang S, Woo JH, Melhem ER (2006) Advanced MR imaging techniques in the diagnosis of intraaxial brain tumors in adults. Radiographics 26:S173–S189
Rajinikanth V, Fernandes SL, Bhushan B, Sunder NR (2018) Segmentation and analysis of brain tumor using Tsallis entropy and regularised level set. In: Proceedings of 2nd international conference on micro-electronics, electromagnetics and telecommunications, pp 313–321
Perkins A, Liu G (2016) Primary brain tumors in adults: diagnosis and treatment. Am Fam Physician 93:211–217
Davis FG, Malmer BS, Aldape K, Barnholtz-Sloan JS, Bondy ML, Brännström T et al (2008) Issues of diagnostic review in brain tumor studies: from the Brain Tumor Epidemiology Consortium. Cancer Epidemiol Prevent Biomarkers 17:484–489
Sharif M, Amin J, Raza M, Yasmin M, Satapathy SC (2020) An integrated design of particle swarm optimization (PSO) with fusion of features for detection of brain tumor. Pattern Recogn Lett 129:150–157
Sharif MI, Li JP, Khan MA, Saleem MA (2020) Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recogn Lett 129:181–189
Thompson G, Mills S, Coope D, O’connor J, Jackson A (2011) Imaging biomarkers of angiogenesis and the microvascular environment in cerebral tumours. Br J Radiol 84:S127–S144
Fernandes SL, Tanik UJ, Rajinikanth V, Karthik KA (2019) "A reliable framework for accurate brain image examination and treatment planning based on early diagnosis support for clinicians. Neural Comput Appl 32:15897–15908
Durand T, Bernier M-O, Léger I, Taillia H, Noël G, Psimaras D et al (2015) Cognitive outcome after radiotherapy in brain tumor. Curr Opin Oncol 27:510–515
DeAngelis LM (2005) Chemotherapy for brain tumors—a new beginning. ed: Mass Medical Soc
Mittal M, Goyal LM, Kaur S, Kaur I, Verma A, Hemanth DJ (2019) Deep learning based enhanced tumor segmentation approach for MR brain images. Appl Soft Comput 78:346–354
Rehman A, Khan MA, Saba T, Mehmood Z, Tariq U, Ayesha N (2020) Microscopic brain tumor detection and classification using 3D CNN and feature selection architecture. Microsc Res Tech 84(1):133–149
Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS (2020) Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recogn Lett 129:293–303
Liaqat A, Khan MA, Shah JH, Sharif M, Yasmin M, Fernandes SL (2018) Automated ulcer and bleeding classification from WCE images using multiple features fusion and selection. J Mech Med Biol 18:1850038
Khan MA, Sarfraz MS, Alhaisoni M, Albesher AA, Wang S, Ashraf I (2020) StomachNet: optimal deep learning features fusion for stomach abnormalities classification. IEEE Access 8:197969–197981
Khan MA, Rubab S, Kashif A, Sharif MI, Muhammad N, Shah JH et al (2020) Lungs cancer classification from CT images: an integrated design of contrast based classical features fusion and selection. Pattern Recogn Lett 129:77–85
Khan MA, Qasim M, Lodhi HMJ, Nazir M, Javed K, Rubab S et al (2020) Automated design for recognition of blood cells diseases from hematopathology using classical features selection and ELM. Microsc Res Tech 84(2):202–216
Khan MA, Khan MA, Ahmed F, Mittal M, Goyal LM, Hemanth DJ et al (2020) Gastrointestinal diseases segmentation and classification based on duo-deep architectures. Pattern Recogn Lett 131:193–204
Hemanth DJ, Anitha J, Mittal M (2018) Diabetic retinopathy diagnosis from retinal images using modified hopfield neural network. J Med Syst 42:247
Acharya UR, Fernandes SL, WeiKoh JE, Ciaccio EJ, Fabell MKM, Tanik UJ et al (2019) Automated detection of Alzheimer’s disease using brain MRI images—a study with various feature extraction techniques. J Med Syst 43:302
Rajinikanth V, Thanaraj KP, Satapathy SC, Fernandes SL, Dey N (2019) Shannon’s entropy and watershed algorithm based technique to inspect ischemic stroke wound. Smart intelligent computing and applications. Springer, pp 23–31
Amin J, Sharif M, Yasmin M, Saba T, Anjum MA, Fernandes SL (2019) A new approach for brain tumor segmentation and classification based on score level fusion using transfer learning. J Med Syst 43:1–16
Amin J, Sharif M, Yasmin M, Fernandes SL (2018) Big data analysis for brain tumor detection: deep convolutional neural networks. Futur Gener Comput Syst 87:290–297
Nazar U, Khan MA, Lali IU, Lin H, Ali H, Ashraf I et al (2020) Review of automated computerized methods for brain tumor segmentation and classification. Curr Med Imaging 16:823–834
Khan MA, Ashraf I, Alhaisoni M, Damaševičius R, Scherer R, Rehman A et al (2020) Multimodal brain tumor classification using deep learning and robust feature selection: a machine learning application for radiologists. Diagnostics 10:565
Huang W, Xue Y, Wu Y (2019) A CAD system for pulmonary nodule prediction based on deep three-dimensional convolutional neural networks and ensemble learning. PLoS ONE 14:e0219369
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using MRI. Pattern Recogn Lett
Raja NSM, Fernandes S, Dey N, Satapathy SC, Rajinikanth V (2018) Contrast enhanced medical MRI evaluation using Tsallis entropy and region growing segmentation. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-018-0854-8
Remeseiro B, Bolon-Canedo V (2019) A review of feature selection methods in medical applications. Comput Biol Med 112:103375
Zahoor S, Lali IU, Khan MA, Javed K, Mehmood W (2020) Breast cancer detection and classification using traditional computer vision techniques: a comprehensive review. Curr Med Imaging 16(10):1187–1200
Ghaffari M, Sowmya A, Oliver R (2019) Automated brain tumor segmentation using multimodal brain scans: a survey based on models submitted to the BraTS 2012–2018 challenges. IEEE Rev Biomed Eng 13:156–168
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, et al (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629
Hussain UN, Khan MA, Lali IU, Javed K, Ashraf I, Tariq J et al (2020) A unified design of ACO and skewness based brain tumor segmentation and classification from MRI scans. J Control Eng Appl Inform 22:43–55
Khan MA, Lali IU, Rehman A, Ishaq M, Sharif M, Saba T et al (2019) Brain tumor detection and classification: a framework of marker-based watershed algorithm and multilevel priority features selection. Microsc Res Tech 82:909–922
Nazir M, Khan MA, Saba T, Rehman A (2019) Brain tumor detection from MRI images using multilevel wavelets. Int Conf Comput Inf Sci (ICCIS) 2019:1–5
Sharif M, Tanvir U, Munir EU, Khan MA, Yasmin M (2018) Brain tumor segmentation and classification by improved binomial thresholding and multi-features selection. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-018-1075-x
Narmatha C, Eljack SM, Tuka AARM, Manimurugan S, Mustafa M (2020) A hybrid fuzzy brain-storm optimization algorithm for the classification of brain tumor MRI images. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02470-5
Rehman A, Naz S, Razzak MI, Akram F, Imran M (2020) A deep learning-based framework for automatic brain tumors classification using transfer learning. Circ Syst Signal Process 39:757–775
Mzoughi H, Njeh I, Wali A, Slima MB, BenHamida A, Mhiri C et al (2020) Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J Digital Imaging 33(4):903–915
Khan MA, Zhang Y-D, Sharif M, Akram T (2021) Pixels to classes: intelligent learning framework for multiclass skin lesion localization and classification. Comput Electr Eng 90:106956
Hussain N, Khan MA, Sharif M, Khan SA, Albesher AA, Saba T et al (2020) A deep neural network and classical features based scheme for objects recognition: an application for machine inspection. Multimed Tools Appl. https://doi.org/10.1007/s11042-020-08852-3
Raza M, Sharif M, Yasmin M, Khan MA, Saba T, Fernandes SL (2018) Appearance based pedestrians’ gender recognition by employing stacked auto encoders in deep learning. Futur Gener Comput Syst 88:28–39
Fernandes SL, Rajinikanth V, Kadry S (2019) A hybrid framework to evaluate breast abnormality using infrared thermal images. IEEE Consumer Electron Mag 8:31–36
Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15:1929–1958
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Zhuang F, Qi Z, Duan K, Xi D, Zhu Y, Zhu H, et al (2020) A comprehensive survey on transfer learning. In: Proceedings of the IEEE
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Krizhevsky A, Sutskever I, Hinton GE (2017) Imagenet classification with deep convolutional neural networks. Commun ACM 60:84–90
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition pp 2818–2826
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group No. (RG-1438-034). The authors would like to thank the Deanship of Scientific Research and RSSU at King Saud University for their technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest in this work and all authors equally work in this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharif, M.I., Khan, M.A., Alhussein, M. et al. A decision support system for multimodal brain tumor classification using deep learning. Complex Intell. Syst. 8, 3007–3020 (2022). https://doi.org/10.1007/s40747-021-00321-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00321-0