Abstract
Invertebrates represent a large proportion of the biomass in aquatic ecosystems, particularly in the ocean. In recent years, invertebrates have been well-consolidated models in evolutionary developmental biology research and, consequently, genomic and transcriptomic sequence databases from a plethora of species across the animal kingdom have become available. This has provided an excellent source of evidence confirming that invertebrates operate endogenous mechanisms for PUFA production. The present paper reviews the current knowledge of gene complement and, where possible, the function of desaturases and elongases with pivotal roles in PUFA biosynthesis of aquatic invertebrates. More specifically, this review covers three major enzyme types, namely ωx desaturases, front-end desaturases and elongases, that have been characterised from species of sponges, cnidarians, molluscs, annelids, crustaceans, rotifers, echinoderms and non-vertebrate chordates (amphioxus and sea squirt). These studies have shown that invertebrates operate alternative and unusual pathways of PUFA biosynthesis involving gene families with complex phylogeny and functional diversity. Consequently, research in this area provides potentially valuable molecular tools in the form of genes that can be used in the biotechnological production of n-3 products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seafood is regarded as an important component of a healthy diet for humans, providing highly digestible protein and micronutrients such as minerals and vitamins. Among all nutrients seafood provides to the human diet, the so-called “omega-3” (or n-3) long-chain (C20–24) polyunsaturated fatty acids (PUFA) have been the most associated with the benefits of seafood consumption by humans. Evidence collected from epidemiological studies, intervention trials and laboratory-based mechanistic studies support beneficial roles of n-3 long-chain PUFA in a variety of human conditions including cardiovascular disease (CVD) and other inflammatory diseases (e.g., rheumatoid arthritis, Crohn’s disease and ulcerative colitis), some types of cancer (e.g., colorectal, breast and prostate) and ensure normal cognitive and visual function during early development (Brouwer et al. 2006).
Compared to terrestrial ecosystems, the marine environment is characterised by the abundance of n-3 long-chain PUFA. Primary production of n-3 long-chain PUFA in the ocean has been historically associated exclusively to single-cell microorganisms occupying the lower trophic levels of the marine food web, including photosynthetic microalgae, heterotrophic protists and bacteria. Higher-trophic-level organisms such as invertebrates or fish were regarded as contributing to “trophic upgrading”, a phenomenon by which PUFA originated from primary producers can be modified as they pass up the food chain (Ahlgren et al. 2009; Brett et al. 2009; Desvilettes and Bec 2009). As described in detail below, a recent study has revealed that multiple invertebrates, many of them representing abundant groups in aquatic ecosystems, possess enzymes that enable them to produce long-chain PUFA de novo and, similarly to single-cell microorganisms, can also contribute to n-3 long-chain PUFA production (Kabeya et al. 2018a).
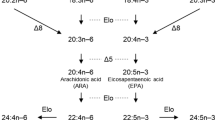
De novo biosynthesis of PUFA by marine microbes can occur via different pathways and comprehensive reviews on this topic include those by Pereira et al. (2003), Uttaro (2006) and Wang et al. (2013). Briefly, microalgae, like all living organisms, have the ability to introduce the first unsaturation into saturated fatty acids (FA; primarily 18:0 but also 16:0) to produce monounsaturated FA such as oleic acid (OA; 18:1n-9) or palmitoleic acid (16:1n-7). The introduction of further double bonds (unsaturations) into OA proceeds via an aerobic pathway involving Δ12 (or ω6) desaturases to produce 18:2n-6 (LA; linoleic acid), which can be subsequently desaturated to 18:3n-3 (ALA; α-linolenic acid) by the action of a Δ15 (or ω3) desaturase (Fig. 1; Guschina and Harwood 2006a). Both Δ12 and Δ15 desaturases are categorised as “methyl-end desaturases” because they introduce the new double bond between an existing unsaturation and the methyl terminus (–CH3) of the FA. However, another type of desaturases called “front-end desaturases”, capable of inserting double bonds between the existing double bonds (typically at Δ9 position) and the carboxyl group (–COOH), and other enzymes known as “elongases”, convert ALA to long-chain PUFA such as 20:5n-3 (EPA; eicosapentaenoic acid) and 22:6n-3 (DHA; docosahexaenoic acid), and LA to 20:4n-6 (ARA; arachidonic acid). Typically, from ALA, the pathway Δ6 desaturase → elongase → Δ5 desaturase enables the production of EPA, with further elongase → Δ4 desaturase enabling DHA biosynthesis (Fig. 1; Guschina and Harwood 2006a). Some species can operate alternative routes for EPA biosynthesis, which involve Δ8 or Δ17 desaturation reactions (Fig. 1) (Guschina and Harwood 2006a). ARA biosynthesis from LA requires the same reactions (Fig. 1). Marine bacteria have been recognised to operate conventional aerobic pathways to biosynthesise PUFA described above (Russell and Nichols 1999) but, additionally, selected species (e.g., Shewanella sp., Vibrio sp.) can also produce PUFA via the anaerobic pathway involving the enzymatic complex polyketide synthase (PKS), which consists of steps adding C2 units and unsaturations (Metz et al. 2001). A similar enzyme complex named “PUFA synthase” reported in the marine protist Schizochytrium contains genes that account for the production of DHA and 22:5n-6 (osbond acid or n-6 docosapentaenoic acid) characteristic of this species (Barclay et al. 1994). There exists evidence suggesting that, as reported above for some bacteria, the anaerobic PUFA synthase pathway appears to coexist with enzymes of the aerobic pathway (Qiu et al. 2001).
Biosynthetic pathways of polyunsaturated fatty acids. Desaturation reactions are denoted with “ω” of “Δ” to refer, respectively, the carbon position at which the incipient double bond (unsaturation) locates within the methyl (ω; green arrows) and front (Δ) ends (orange arrows) of fatty acyl chains. Elongation reactions are denoted with “Elo” (blue arrows). β-ox β-oxidation, OA oleic aci, LA linoleic acid, ALA α-linolenic acid, ARA arachidonic acid, EPA eicosapentaenoic acid, DHA docosahexaenoic acid (colour figure online)
Like all vertebrates, fish cannot biosynthesise PUFA de novo since they lack Δ12 and Δ15 desaturase activities required to produce LA and ALA from OA (Fig. 1). However, fish do possess fatty acyl (front-end) desaturases (Fads) and elongation of very long-chain fatty acid (Elovl) proteins (commonly known as “elongases”) (Castro et al. 2016), both with pivotal roles in the production of long-chain (C20–24) PUFA from the C18 PUFA precursors LA and ALA (Fig. 1). In contrast to other vertebrates which have two types of Fads desaturases, namely Fads1 (∆5 desaturase) and Fads2 (∆6 desaturase) (Guillou et al. 2010), teleost fish appear to have lost fads1 during evolution and thus have a varied number of fads2 genes (Castro et al. 2012, 2016). To date, only one exception to this pattern has been reported in the Japanese eel Anguilla japonica, a basal teleost that possesses a Fads1 with ∆5 desaturase capacity (Lopes-Marques et al. 2018). More commonly though, teleostei Fads2 is functionalised during expansion of teleosts and Fads2 with Δ6 (Zheng et al. 2004, 2009; González-Rovira et al. 2009; Mohd-Yusof et al. 2010; Monroig et al. 2010a, 2013a; Kabeya et al. 2018b), dual Δ6 and Δ5 (Hastings et al. 2001; Tanomman et al. 2013; Fonseca-Madrigal et al. 2014; Kuah et al. 2016; Oboh et al. 2016; Ferraz et al. 2018), Δ5 (Abdul Hamid et al. 2016) and Δ4 (Li et al. 2010; Morais et al. 2012; Fonseca-Madrigal et al. 2014; Oboh et al. 2017a) desaturase activities have been identified. Like mammalian FADS2 (Park et al. 2009), teleost Fads2 typically show Δ8 desaturase activity (Monroig et al. 2011a), enabling the initiation of the long-chain PUFA biosynthetic pathways via the so-called “Δ8 pathway”, an alternative route to the “Δ6 pathway” (Fig. 1) (Castro et al. 2016).
Elovl enzymes catalyse the rate-limiting step (condensation) in the elongation pathway that results in an extension of the FA by two carbons (Leonard et al. 2004). In vertebrates, three members of the Elovl family, namely Elovl2, Elovl4 and Elovl5, have key roles in PUFA biosynthesis (Leonard et al. 2004; Jakobsson et al. 2006). In fish, Elovl5 enzymes can efficiently elongate C18 and C20 PUFA, with remarkably lower elongase capacity towards C22 substrates (Castro et al. 2016). Elovl2, an enzyme with C20 and C22 PUFA as preferred elongation substrates, exists in some fish (Monroig et al. 2009; Morais et al. 2009; Oboh et al. 2016; Ferraz et al. 2018), but it was lost during expansion of teleosts and is absent in recently emerged teleost lineages (Castro et al. 2016). Teleost Elovl4, along with its role in the biosynthesis of very long-chain (> C24) PUFA (Monroig et al. 2010b), can elongate C22 PUFA like Elovl2 and thus has been hypothesised to somewhat compensate for the loss of Elovl2 in some teleosts (Monroig et al. 2011b; Kabeya et al. 2015; Jin et al. 2017; Oboh et al. 2017b).
From the above studies, it is now well understood that the repertoire and function of genes encoding Fads and Elovl varies among fish species, thus determining the extent to which each species can biosynthesise C20–24 PUFA such as EPA, ARA and DHA from C18 PUFA. EPA, ARA and DHA play physiologically important roles in vertebrates, including fish, and this has driven the interest to understand the capacity that each species has to desaturate and elongate C18 PUFA contained in many vegetable oils extensively used in aquafeed to replace fish oil (Turchini et al. 2010). Comprehensive reviews on this topic have been published recently (Tocher 2015; Castro et al. 2016; Kabeya et al. 2017a; Monroig et al. 2018).
Unlike single-celled organisms and fish, the pathways of PUFA biosynthesis in invertebrates have been barely investigated (Monroig et al. 2013b). However, ever-increasing genomic data are becoming available for many invertebrate species. This offers a unique opportunity to mine their genomes for genes encoding key enzymes of PUFA biosynthesis in invertebrates and helps to establish which enzymatic activities derive from the animal itself and which from microorganisms that can potentially produce PUFA and inhabit the invertebrate hosts (Jøstensen and Landfald 1997). From a functional standpoint, there is considerable activity around the globe in biotechnology companies to produce new sources of n-3 long-chain PUFA that will alleviate the pressure on fish stocks to guarantee an increasing demand (Tocher 2015). Clearly, marine invertebrates represent an interesting and barely explored source of desaturase and elongase enzymes, potentially with novel functionalities, amenable for biotechnological production of n-3 products (Xue et al. 2013; Hamilton et al. 2016). In this context, this review focuses on recent research into the molecular and biochemical mechanisms of PUFA biosynthesis in invertebrates within groups with particular importance in aquatic ecosystems including sponges, cnidarians, nematodes, rotifers, molluscs, polychaetes, echinoderms, crustaceans and non-vertebrate chordates (tunicates and amphioxus). Particular attention is paid to studies reporting on functional data of desaturase and elongase enzymes involved in the pathways of de novo and trophic upgrading of PUFA in invertebrates. As explained below, the phylogeny of animal desaturase and elongase genes with roles in PUFA biosynthesis is complex and, in most cases, has not yet been resolved. For simplicity, we have herein established three categories of enzymes (methyl-end desaturases, front-end desaturases and Elovl enzymes) based on their substrate specificity rather than their phylogeny. Such classification complexity is particularly relevant for desaturases since, within each of the methyl-end and front-end desaturase categories, we have identified gene lineages with clearly distinct origins despite them operating rather similar regioselectivity modes.
Methyl-end desaturases
Methyl-end desaturases (also known as “ωx desaturases”) have been traditionally believed to be absent in most animals, with some exceptions, including the terrestrial nematode Caenorhabditis elegans (Spychalla et al. 1997; Peyou-Ndi et al. 2000; Zhou et al. 2011). Additionally, a number of arthropods, mainly hexapods, have shown the capability of de novo biosynthesis of PUFA but, with the exception of the whitefly Bemisia argentifolii (Buckner and Hagen 2003), such arthropod enzymatic capacity has been mostly limited to biosynthesis of LA but not ALA (Malcicka et al. 2017). Further in vivo evidence for de novo production of LA and/or ALA has been shown in terrestrial molluscs (Weinert et al. 1993), aquatic species of polychaetes (Pairohakul 2013) and crustaceans (Farkas et al. 1981). Biosynthesis of ALA from LA has been reported in Artemia sp. (Schauer and Simpson 1985; Ito and Simpson 1996), but contribution from PUFA-biosynthesising microbes could not be totally excluded.
Functional characterisation through expression of genome-anchored ωx desaturases in heterologous systems such as yeast or, less commonly, plants, allows us to establish which specific genes enable the de novo PUFA biosynthesis in invertebrates. The first demonstration of the existence of an ωx desaturase in animals was the C. elegans Fat-1 (Spychalla et al. 1997). Its expression in Arabidopsis revealed that Fat-1 could be categorised as an “ω3 desaturase” since it showed Δ15 desaturase activity towards LA to produce ALA, and Δ17 desaturase activity towards ARA, which was converted to EPA (Spychalla et al. 1997). A further ωx desaturase from C. elegans, namely Fat-2, was characterised in yeast Saccharomyces cerevisiae, first as a Δ12 desaturase (Peyou-Ndi et al. 2000) and subsequently reported to have also Δ15 desaturase activity (Zhou et al. 2011). Further animal ωx desaturases with Δ12 desaturase activity have also been characterised in the two insects (Arthropoda) Acheta domesticus and Tribolium castaneum (Zhou et al. 2008). Interestingly, despite their Δ12 desaturase capacity, the insect desaturases have a different evolutionary origin compared to the C. elegans ωx desaturases and those recently found in multiple invertebrates (Kabeya et al. 2018a), suggesting functional convergence among animal desaturases.
Indeed, Kabeya et al. (2018a) have demonstrated that ωx desaturase genes enabling PUFA biosynthesis de novo are widespread among invertebrates since a total of 121 ωx desaturase sequences were found in 80 species within Cnidaria, Nematoda, Annelida, Mollusca, Rotifera and Arthropoda. No ωx desaturase-like sequence was found in genomes from Porifera, Ctenophora, Priapulida and Brachiopoda, or Deuterostoma including Echinodermata, Hemichordata or Chordata. Interestingly, the animal ωx desaturase genes were not derived from a single monophyletic origin, but rather separated into three well-supported distinct clades in the molecular phylogenetic analysis. The main three clades (clades 1–3) grouping most of animal ωx desaturases included sequences from nematodes (clade 1), cnidarians (clade 2) and a combination of sequences from rotifers, molluscs, annelids and arthropods (clade 3) (Kabeya et al. 2018a). The existence of three major clades suggests that the animal ωx desaturases have distinct evolutionary origins. Further instances of animal ωx desaturases outside the three major clades were found in some terrestrial arthropods, namely Hemiptera Bemisia tabaci, Orthoptera Locusta migratoria and Collembola Sminthurus viridis (Kabeya et al. 2018a). Indeed, the B. tabaci ωx desaturases were demonstrated to be cases of horizontal gene transfer (HGT), but it does not appear to be the only case (Spychalla et al. 1997; Peyou-Ndi et al. 2000; Zhou et al. 2011). Thus, HGT is the most plausible explanation for the general phylogenetic pattern of animal ωy desaturases, but instances of gene loss, gene duplication and convergent evolution have clearly impacted the animal ωy desaturase genes (Kabeya et al. 2018a). In the sections below, we summarise the functional analysis of the ωx desaturases within animal representatives from clades 1–3 as defined by Kabeya et al. (2018a), which encompass the majority of aquatic invertebrates in which these novel desaturases have been identified.
Nematodes
As mentioned above, clade 1, consisting exclusively of nematode ωx desaturase sequences, included the functionally characterised Fat-1 (ω3 desaturase) and Fat-2 (Δ12/Δ15 desaturase) from C. elegans (Spychalla et al. 1997; Peyou-Ndi et al. 2000; Zhou et al. 2011). Due to the limited number of nematode genomic and transcriptomic databases available to date, only parasitic or terrestrial free-living species of nematodes were identified to possess ωx desaturases. However, it is likely that aquatic nematodes, widely distributed in the marine environment (Meldal et al. 2007), also possess ωx desaturase genes and thus can play a pivotal role for the input of n-3 PUFA to the marine ecosystem. While not strictly aquatic, the free-living nematode Panagrellus redivivus, often used to feed early life-cycle stages of fish (Brüggemann 2012), showed Δ12 desaturase activity since 14C-labelled 18:1n-9 was converted into 18:2n-6 (LA) in in vivo experiments (Schlechtriem et al. 2004). Based on the relatively high contents of n-3 FA in the experimental nematodes, the presence of endogenous Δ15 desaturase activity in P. redivivus was also postulated given that these could not be explained from dietary input (Schlechtriem et al. 2004).
Cnidarians
Clade 2 comprised ωx desaturase sequences exclusively from cnidarians, more specifically from stony corals (order Scleractinia), coral anemones (Corallimorpharia), sea anemones (Actiniaria) and zoanthids (Zoantharia) (Kabeya et al. 2018a). It is worth noting that ωx desaturase-like sequence cannot be found in Hydrozoa, in spite of the existence of well-annotated genomic assembly for Hydra vulgaris (Chapman et al. 2010). Within clade 2, some species possess two distinct ωx desaturase genes, each forming two distinct gene lineages suggesting the occurrence of gene duplication early in the evolution of Cnidaria. This is the case in Acropora millepora (stony coral) in which one ωx desaturase was functionally characterised as a Δ12 desaturase, whereas a second ωx desaturase was characterised as an ω3 desaturase (Kabeya et al. 2018a). The latter can desaturate a series of ω6 substrates, namely C18 (LA and 18:3n-6), C20 (20:2n-6, 20:3n-6 and ARA) and C22 (22:4n-6) FA, confirming Δ15, Δ17 and Δ19 desaturase activities, respectively. Coexistence of those three desaturase activities within a single enzyme is a rather uncommon feature among previously characterised ω3 desaturases (Wang et al. 2013). Beyond the potential implications for the n-3 oil production industry, these results have important ecological implications since they suggest that coral reefs, ecosystems dominated by stony corals and coral anemones with ωx desaturases similar to those described in A. millepora, arise as important production sites of ω3 PUFA at a global scale. While endosymbionts such as Symbiodinium sp. have been suggested to provide essential lipids including long-chain PUFA to their coral hosts (Imbs et al. 2014), the results by Kabeya et al. (2018a) demonstrate that corals themselves have endogenous PUFA production capacity to satisfy, at least in part, their physiological demands.
Other invertebrates (rotifers, molluscs, annelids and arthropods)
The third well-supported animal ωx desaturase gene lineage (clade 3) included sequences from rotifers, molluscs, annelids and arthropods (Kabeya et al. 2018a). Like observed in cnidarians, two distinct ωx desaturases were identified for a substantial number of species within these phyla but, in contrast to cnidarian genes, they did not form two distinct clades in the phylogenetic tree (Kabeya et al. 2018a). Intriguingly, the distribution of ωx desaturase genes within particular taxonomic groups in this clade is rather cryptic. In rotifers, the genes can only be found in bdelloid rotifers including Adineta vaga and several species from genus Rotaria, but not in monogonont rotifers like Brachionus. A similar case applies to bivalve molluscs, since most of marine species such as Pacific oyster Crassostrea gigas and Mediterranean mussel Mytilus galloprovincialis (both from order Ostreoida) appear to lack ωx desaturase genes, while the gene can be found in Elliptio complanata, a freshwater mussel that belongs to the order Unionoida (Kabeya et al. 2018a). Arguably, the most extreme case was observed in crustaceans, whereby ωx desaturase genes were exclusively found in specific orders of copepods, namely Siphonostomatoida, Cyclopoida and Harpacticoida, but not Calanoida copepods. The marine pelagic ecosystem is extremely rich in n-3 PUFA produced by photosynthetic microalgae and therefore it can be hypothesised that the evolutionary and ecological pressure to retain ωx desaturases would be low in planktonic copepods (e.g., calanoids) and high in benthic copepods (e.g., harpacticoids).
The functions of ωx desaturases from clade 3 have been characterised in representative species of rotifers (A. vaga), molluscs (Patella vulgata), annelids (Riftia pachyptila and Platynereis dumerilii) and arthropods (Lepeophtheirus salmonis) (Liu et al. 2017a; Kabeya et al. 2018a). With the exception of A. vaga and R. pachyptila, a fairly conserved pattern was observed in P. vulgata, P. dumerilii and L. salmonis, with existence of two distinct genes with complementary substrate specificities, more specifically a Δ12 desaturase and an ω3 desaturase with Δ15, Δ17 and Δ19 desaturase activities (Fig. 1). Such enzymatic activities are largely consistent with those collected from in vivo studies in the rugworm Alitta (Nereis) virens, a nereid polychaete like P. dumerilii, which was capable of de novo biosynthesis of ARA and EPA when fed pellets supplemented with 13C-labelled palmitic acid (16:0) (Pairohakul 2013). Instead of separated Δ12 and ω3 desaturases as described for P. vulgata, P. dumerilii and L. salmonis, a different pattern of enzymatic capabilities was found in ωx desaturases from the rotifer A. vaga. Among the three ωx desaturases found in A. vaga, the two ωx desaturases characterised by Kabeya et al. (2018a) exhibited rather similar activities since both possessed simultaneously Δ12, Δ15 and Δ17 desaturase activities. Unlike other ω3 desaturases found in cnidarian, molluscs, annelids and arthropods, no Δ19 desaturase activity was detected in the two A. vaga ωx desaturases studied (Kabeya et al. 2018a). Interestingly, the sole ωx desaturase characterised from the giant tube worm R. pachyptila (Liu et al. 2017a) showed Δ15 desaturase capacity but not Δ17 and Δ19 activities (Liu et al. 2017a). Since two genes could be identified in Lamellibrachia satsuma (Kabeya et al. 2018a), a closely related polychaete species to R. pachyptila, it is reasonable to expect that R. pachyptila possesses another ωx desaturase gene, probably with Δ12 desaturase that complements the Δ15 desaturase activity of the former ωx desaturase (Liu et al. 2017a) and thus enables the de novo biosynthetic pathway of ALA in this species. Such an endogenous ability to produce physiologically important PUFA would represent an important ecological adaptation for the giant tubeworm R. pachyptila to environments with little input of these compounds via major primary producers such as photosynthetic microalgae (Liu et al. 2017a). Similarly, the aquatic invertebrate communities largely composed of annelids, molluscs and arthropods dominate benthic ecosystems, and therefore possession of ωx desaturases appears as an advantageous trait to counteract the naturally poor availability of n-3 PUFA in such environments (Vonk et al. 2016).
Front-end desaturases
Sponges
FA from sponges, often referred to as “demospongic acids” since they have been described primarily in Demospongiae species, have unusual structures including very long-chain (up to C34) and high degrees of unsaturation (up to six double bonds) (Barnathan 2009; Kornprobst and Barnathan 2010). Importantly, demospongic acids also have a characteristic pattern of unsaturation consisting of Δ5,9 double bonds (or elongation products of them) resulting in a particular type of PUFA called “non-methylene interrupted” (NMI) FA. The desaturation pathway to produce NMI FA has been proposed from in vivo assays using radiolabelled FA substrates and analytical evidence from FA composition (Barnathan 2009; Kornprobst and Barnathan 2010). Briefly, the first unsaturation can be introduced at either Δ5 or Δ9 positions, and a subsequent unsaturation is inserted at either side of the pre-existing one to form a second double bond at positions Δ9 or Δ5. In addition to dienes with Δ5,9 or elongation-derived combinations such as Δ7,11-, Δ9,13-, Δ11,15-, Δ17,21-, Δ19,23- Δ21,25- and Δ23,27- (Kornprobst and Barnathan 2010), demospongic acids also include trienes like 26:3Δ5,9,19 described in the freshwater sponge Ephydatia fluviatilis (Hahn et al. 1988, 1989). The double bond at the Δ19 position of 26:3Δ5,9,19 would result from the elongation of the monounsaturated FA 16:1Δ9 to 26:1Δ19, and then sequential desaturation of Δ5 and Δ9 positions via the abovementioned pathway. In addition to the above studies, FA having desaturations at ∆6 and ∆11 positions were identified in the sponge Euryspongia rosea (Carballeira and Maldonado 1989), implying the existence of a ∆6 desaturation pathway. These results suggest that sponges possess, in addition to Δ9 desaturases, front-end desaturases with Δ5 and ∆6 desaturase capacities, although, to the best of our knowledge, no molecular evidence establishing the role of a specific fatty acyl desaturase from sponges has been published. Nevertheless, the well-annotated genomic assembly of the demosponge Amphimedon queenslandica has been released in the NCBI (accession no. GCA_000090795.1; Srivastava et al. 2010) and, despite annotations having been generated by computational gene prediction, multiple desaturase-like sequences can be found in this species (Gold et al. 2017). Functional characterisation of sponge front-end desaturases will help to elucidate the pathways of biosynthesis of NMI FA and other PUFA found in sponges, and clarify the contributions of endosymbionts that are often associated with these animals.
Cnidarians
Endogenous production of PUFA in some species of anthozoans (corals and sea anemones) and scyphozoans (jellyfish) are partly explained by contributions from the endosymbiotic dinoflagellates Symbiodinium (or “zooxanthellae”) (Díaz-Almeyda et al. 2011), which thus help to partly satisfy the FA requirements of the host (Papina et al. 2003; Imbs et al. 2014). Indeed, FA translocation from endosymbionts to the host has been described to occur in both anthozoans (Muscatine et al. 1994) and scyphozoans (Mortillaro et al. 2009). Imbs et al. (2010) separately analysed the profiles of PUFA in zooxanthellae, polyp tissue and intact colonies in Sinularia sp. (Octocorallia) and Acropora sp. (Hexacorallia). The results suggested that n-3 PUFA, namely 18:4n-3, EPA, 22:5n-3 and DHA, were mainly produced by the zooxanthellae, but, importantly, some n-6 PUFA (20:3n-6, ARA, and 22:4n-6) were biosynthesised by the polyp, confirming that the host can actively contribute to endogenous production of PUFA. The characteristic C24 PUFA (24:5n-6 and 24:6n-3), which can be used for chemotaxonomic markers of soft corals (Svetashev and Vysotskii 1998), were identified in Sinularia sp. polyps regardless of the presence or absence of zooxanthellae, suggesting that their biosynthesis occurs in the coral polyps (Imbs et al. 2010). The authors proposed the action of a Δ6 desaturase in the production of 16:2n-7 by Sinularia sp. and 18:3n-6 by Acropora sp. (Imbs et al. 2010).
Surm et al. (2018) recently examined the repertoire of front-end desaturases (termed “Fad”) and Elovl genes in the phylum Cnidaria, in which genomes from representatives of a range of cnidarian taxa (H. vulgaris, Acropora digitifera, Nematostella vectensis and Exaiptasia pallida) have been interrogated. Moreover, the expression of cnidarian Fad (and Elovl) genes was further confirmed through extensive searches against transcriptome assemblies within six actiniarian species (Surm et al. 2018). The copy number of Fad-like genes varied among species, with species like H. vulgaris and N. vectensis having one gene, while three copies exist in E. pallida. The Cnidaria Fad genes formed two distinct clades, one including functionally characterised Δ6 desaturases from the green plant Borago officinalis and another clade including the Δ5 desaturase from the fungus Mortierella alpina (Surm et al. 2018). Unfortunately, this interesting study did not provide functional data and thus it remains unknown whether the cnidarian Fad retrieved in silico are indeed front-end desaturases with similar functions as their plant or fungal sister sequences. More clearly though, Fad genes hypothesised to play roles in PUFA biosynthesis in cnidarians cannot be regarded as orthologues of vertebrate front-end desaturases (i.e., Fads), since the latter formed a separate clade from the two Cnidaria clades described above (Surm et al. 2018). These results emphasise the diversity of desaturase enzymes with putative roles in PUFA biosynthesis in animals.
Molluscs
The long-chain PUFA biosynthetic capability of molluscs has been extensively investigated in comparison to other invertebrate groups, partly due to their commercial importance and broad interest as a healthy n-3 long-chain PUFA source in the human’s food basket (Monroig et al. 2013b). The FA profiles of several taxonomic groups of molluscs were comprehensively reviewed by Joseph (1982, 1989) and many studies reported on the capacity for PUFA biosynthesis in cephalopods (Reis et al. 2014, 2016), gastropods (van der Horst 1973, 1974; Weinert et al. 1993; Zhu et al. 1994) and bivalves (De Moreno et al. 1976; Pirini et al. 2007; Waldock and Holland 1984; Zhukova 1986, 1991). One major conclusion from the above studies is that the ability of molluscs for PUFA biosynthesis varies among species as a result of existing enzyme types (e.g., ωx desaturases) and their specific enzymatic specificities.
Cloning and functional characterisation of front-end desaturases involved in the PUFA biosynthetic pathway have been carried out in several species of molluscs. A pioneer study on the common octopus Octopus vulgaris (Monroig et al. 2012a) demonstrated that this species has a front-end desaturase gene with Δ5 desaturase activity that enables the production of ARA and EPA from 20:3n-6 and 20:4n-3, respectively. Subsequently, Δ5 desaturases have been also identified in other cephalopods like Sepia officinalis (Monroig et al. 2016a), gastropods like Haliotis discus hannai (Li et al. 2013), and bivalves such as Chlamys nobilis (Liu et al. 2014a) and Sinonovacula constricta (Ran et al. 2018). Duplicate genes encoding Δ5 desaturases appear to exist in H. discus hannai and S. constricta (Li et al. 2013; Ran et al. 2018). All the front-end desaturases isolated from molluscs possess typical features conserved among animal front-end desaturases, namely a cytochrome b5-like domain with heme-binding motif (HPGG), three histidine boxes consisting of HXXXH, HXXHH and QXXHH, and several membrane-spanning domains. Molecular phylogenetic analysis confirmed that front-end desaturase genes from molluscs can be separated in two distinct well–supported clades named clade A and B (Surm et al. 2015). Clade A included all Δ5 desaturase sequences characterised from O. vulgaris, S. officinalis, H. discus hannai and C. nobilis, and hence it is reasonable to expect that these desaturases (thereafter referred as “FadsA”) are all Δ5 desaturases. In addition, to catalyse the Δ5 desaturation leading to ARA and EPA production (Fig. 1), FadsA from cephalopods and scallops appear to be involved in the biosynthesis of NMI FA (Barnathan 2009), as confirmed by 20:2n-6 (20:2Δ11,14) and 20:3n-3 (20:3Δ11,14,17) being Δ5 desaturated to their corresponding NMI FA 20:3Δ5,11,14 and 20:4Δ5,11,14,17, respectively (Monroig et al. 2012a, 2016a; Liu et al. 2014a). Other NMI FA, namely 20:2∆5,11 and 20:2∆7,13, were also detected in lipids from O. vulgaris, suggesting that FadsA produce NMI FA in coordination with other enzymes such as stearoyl-CoA desaturase (Δ9) (Monroig et al. 2017) and elongases (Monroig et al. 2012b). Moreover, mollusc FadsA also show Δ5 desaturase activity towards saturated FA and are able to desaturate, for instance, 18:0–18:1Δ5 (18:1n-13), a compound that has been found abundant in molluscs such as Littorina littorea and Lunatia triseriata (Joseph 1982).
Non FadsA desaturases have been characterised from the bivalves C. nobilis and S. constricta (Liu et al. 2014b; Ran et al. 2018). More specifically, a desaturase from C. nobilis was characterised as a Δ8 desaturase (Liu et al. 2014b), whereas S. constricta has a Δ6 desaturase (Ran et al. 2018). Despite the respective phylogenetic analyses being somewhat unclear, these desaturases appear to belong to clade B from Surm’s study (Surm et al. 2015; thereafter termed “FadsB”). Unlike FadsA which includes exclusively Δ5 desaturases, FadsB appears to be a more heterogeneous group since it includes both Δ6 (Ran et al. 2018) and Δ8 (Liu et al. 2014b) desaturases. Possessing two front-end desaturases confers species like C. nobilis or S. constricta the ability to perform all desaturation reactions required for biosynthesis of ARA and EPA from LA and ALA, respectively, but operating different pathways in each case (Fig. 1). For C. nobilis, ARA and EPA biosynthesis can proceed via the so-called “Δ8 pathway”, which comprises the initial elongation of LA and ALA to 20:2n-6 and 20:3n-3, respectively, a subsequent Δ8 desaturation to 20:3n-6 and 20:4n-3, and a final Δ5 desaturation to ARA and EPA (Fig. 1). For S. constricta, ARA and EPA biosynthesis would be achieved via the so-called “Δ6 pathway”, which consists of initial Δ6 desaturations of LA and ALA to 18:3n-6 and 18:4n-3, respectively, subsequent elongations to 20:3n-6 and 20:4n-3, and final Δ5 desaturation to ARA and EPA (Fig. 1). Such ability for ARA and EPA biosynthesis appears to be absent in species like O. vulgaris, which have a Δ5 desaturase (FadsA) as the sole putative front-end desaturase existing in their genome (Albertin et al. 2015). These results imply that ARA and EPA are dietary essential for O. vulgaris, a condition that applies to DHA too since, in addition to Δ6 desaturation capacity, lack of Δ4 desaturases also precludes its biosynthesis (Monroig et al. 2012a, 2017).
Annelids
In contrast to sponges (Gold et al. 2017), cnidarians (Surm et al. 2018) and molluscs (Surm et al. 2015), a comprehensive description of front-end desaturase and elongase genes within annelids is missing. However, there is a strong body of evidence showing that active PUFA-biosynthesising machineries involving both front-end desaturase and elongase enzymes exist in annelids, particularly polychatetes (Pond et al. 2002), such as the lugworm Arenicola marina (Olive et al. 2009; Pairohakul 2013), the ragworm Alitta virens (Pairohakul 2013) and the clamworm Perinereis aibuhitensis (Lv et al. 2016). A study by Olive et al. (2009) investigated the PUFA-biosynthesising capacity of A. marina comparing the FA composition of worms grown in mesocosm systems supplemented with brewer’s yeast (i.e., lacking PUFA) under light presence and absence conditions. The potential input from PUFA-biosynthesising bacteria was also studied, but, while their presence could not be totally ruled out, their contribution to the observed enzymatic activities appeared to be negligible. It was concluded that a net production of long-chain PUFA within the worm was evidenced by the presence of metabolic products that were not present in the system, those including the ∆8 desaturation product 20:3n-6, and its resulting ∆5 desaturation product ARA (20:4n-6) (Olive et al. 2009). Therefore, annelids are able to operate the ∆8 pathway for ARA biosynthesis (Fig. 1), but the authors suggested that EPA biosynthesis would be achieved by a ∆17 desaturase from ARA. While it could not be demonstrated then, it is now known that polychaetes do possess ωx desaturases with ∆17 activities that catalyse the conversion of ARA to EPA (Kabeya et al. 2018a).
Crustaceans
Evidence suggesting the existence of front-end desaturases in crustaceans has been primarily collected from investigations on zooplanktons and decapods, due to their ecological importance and economical interest in aquaculture (Morris and Sargent 1973; Kanazawa et al. 1979; Kayama and Hirata 1984; Mourente 1996; De Troch et al. 2012). For example, a study using 14C-labelled LA and ALA demonstrated that Δ6 desaturation converted these substrates into 18:3n-6 and 18:4n-3, respectively, in the calanoid copepod Paracalanus parvus (Moreno et al. 1979). Other studies on Daphnia magna, calyptopus larvae of the Antarctic krill Euphausia superba, and several copepods (calanoids and cyclopoids) further suggested the presence of front-end desaturases (Bell et al. 2007; Farkas et al. 1981). Interestingly, the above studies demonstrated the bioconversion of EPA to DHA in the calanoids Calanus finmarchicus and Drepanopus forcipatus, the cyclopoids Cyclops strenuus and Antarctic krill, but not in D. magna. The presence of front-end desaturases was also suggested in Artemia, since radiolabelled ALA was metabolised to DHA in 15-day old Artemia, when DHA was not present in the diet (Schauer and Simpson 1985).
To the best of our knowledge, no gene-encoding desaturase enzyme that can be unequivocally categorised as front-end desaturase has been reported in crustaceans. In some species of decapod crustaceans, namely Chinese mitten crab Eriocheir sinensis (Yang et al. 2013), the mud crab Scylla paramamosain (Lin et al. 2017), Pacific white shrimp Litopenaeus vannamei (Chen et al. 2017) and Australian red claw crayfish Cherax quadricarinatus (Wu et al. 2018), desaturase genes referred to as “front-end desaturases” have been cloned but not functionally characterised. These desaturase-like sequences have some typical front-end desaturase features including a cytochrome b5-like domain with a heme-binding motif (HPGG) and multiple membrane-spanning regions. Importantly, in the crustacean desaturase, while containing three histidine boxes (Sperling et al. 2003), the amino acid sequence of the third histidine box is HXXHH instead of QXXHH as commonly found in front-end desaturases (Hashimoto et al. 2008; Meesapyodsuk and Qiu 2012). In agreement, the phylogenetic analyses showed that the decapod desaturases clustered distantly from functionally characterised front-end desaturases from other invertebrates such as molluscs and echinoderms (Yang et al. 2013; Lin et al. 2017; Chen et al. 2017; Wu et al. 2018), but rather formed a distinct clade comprising exclusively non-functionally characterised sequences from decapods. As described below, some of these desaturases can be nutritionally regulated in a similar manner as vertebrate Fads are (Monroig et al. 2018). However, their categorisation as front-end desaturases and their substrate specificity as Δ6 desaturases are speculative without adequate phylogenetic and functional characterisation.
Echinoderms
The functions of enzymes involved in PUFA biosynthesis have been barely studied in echinoderms, but front-end desaturases from the sea urchin Paracentrotus lividus (Kabeya et al. 2017b) and the sea cucumber Apostichopus japonicus (Liu et al. 2017a) have been recently characterised. Kabeya et al. (2017b) characterised the functions of the three desaturases identified from P. lividus. Two desaturases showed Δ8 desaturase activity and enabled desaturation of 20:2n-6 and 20:3n-3 to 20:3n-6 and 20:4n-3, respectively. A third desaturase was found to have Δ5 activity towards 20:3n-6 and 20:4n-3, which were converted to ARA and EPA, respectively (Kabeya et al. 2017b). As described above in other invertebrates (C. nobilis and A. marina), P. lividus can operate the Δ8 pathway to biosynthesise ARA and EPA from LA and ALA, respectively (Fig. 1), provided PUFA elongases exist in this species. The P. lividus Δ5 desaturase also had the capacity to produce NMI FA, namely 20:3Δ5,11,14 and 20:4Δ5,11,14,17 from 20:2n-6 and 20:3n-3, respectively (Kabeya et al. 2017b). These results are consistent with the presence of NMI FA in other sea urchin species (Takagi et al. 1986; Liyana-Pathirana et al. 2002). With regards to the sea cucumber A. japonicus, a front-end desaturase was characterised as a Δ6 desaturases since it converted LA and ALA into 18:3n-6 and 18:4n-3, respectively (Liu et al. 2017a).
Such differences in substrate specificities between the P. lividus and A. japonicus desaturases are consistent with these proteins having different evolutionary origins. On one hand, the P. lividus Δ5 desaturase clustered with other Δ5 desaturases from molluscs (i.e., FadsA), as well as putative desaturases from other sea urchins (Strongylocentrotus purpuratus and Lytechinus variegatus) and other echinoderm species of sea lilies (Oxycomanthus japonicus), sea cucumbers (Sclerodactyla briareus) and starfish (Patiria miniata) (Kabeya et al. 2017b). Importantly, Kabeya et al. (2017b) showed that FadsA comprises front-end desaturases with Δ5 desaturase activities that are present not only in molluscs and echinoderms, but also annelids (e.g., Capitella teleta) and hemichordates (Saccoglossus kowalevskii). Moreover, the two Δ8 desaturases from P. lividus clustered with non-FadsA desaturases from S. purpuratus and L. variegatus forming a sea urchin-specific clade termed as “FadsC” by Kabeya et al. (2017b). Finally, the Δ6 desaturase from A. japonicus has a distinct origin since it does not belong to either FadsA or FadsB clusters (Liu et al. 2017a). Irrespective of their functions, the above-described echinoderm desaturases possess all characteristic features of front-end desaturases including the third histidine box as QXXHH. However, their relationship with Fads, the vertebrate front-end desaturases, remains to be clarified.
Elovl enzymes
Sponges
As noted above, lipids from demosponges are characterised by the presence of ∆5,9 NMI FA with chain lengths up to C34. While the atypical desaturation pattern can be accounted for the presence of ∆9 and ∆5 desaturases (see above), the unusually long acyl chain of sponge FA suggests the presence of active FA elongation systems. The study on A. queenslandica by Gold et al. (2017) identified several copies of Elovl-like genes. In addition to Elovl3 and Elovl6, elongases not involved in PUFA elongation (Jakobsson et al. 2006), A. queenslandica possess a single copy of Elovl1/2/4/5/7 (NCBI accession no.: XP_011405756.2), representing the ancestor protein existing before gene duplication events lead to other Elovl gene families such as Elovl2, Elovl4 and Elovl5, with well-established roles in PUFA biosynthesis (Jakobsson et al. 2006; Guillou et al. 2010). Unfortunately the function of the sponge Elovl1/2/4/5/7 remains unknown, but it is reasonable to speculate that it shares enzymatic activities enabling elongation of saturated FA (characteristic of vertebrate Elovl1 and Elovl7) and PUFA (characteristic of vertebrate Elovl2, Elovl4 and Elovl5).
Cnidarians
From interrogation of genomes from several cnidarian species, Surm et al. (2018) concluded that the number of Elovl genes varies among taxa. Thus, one single copy of Elovl genes were found in H. vulgaris and N. vectensis, three in A. digitifera and four in E. pallida, the latter also being the species with the highest desaturase-like genes (Surm et al. 2018). Additionally, the analysis of the Actiniaria transcriptomes revealed that multiple Elovl genes (2–5) resulted via gene duplication, with copy numbers varying within superfamilies (Actinioidea vs. Metridioidea) and among species within these taxa (Surm et al. 2018). The phylogenetic analysis showed that cnidarians possess Elovl4 elongases. However, it could not be clearly established which cnidarian Elovl-like families are more closely related to vertebrate Elovl2 and Elovl5, elongases with known roles in PUFA elongation (Jakobsson et al. 2006; Guillou et al. 2010). Among the potential candidates, two novel cnidarian Elovl clades were identified, namely ElovlA and ElovlB (Surm et al. 2018). The functions of elongases from the two novel Elovl subclades is currently unknown, but lack of 22:5n-3 and DHA in tissues of Actinia tenebrosa might suggest that none of them were able to elongate PUFA substrates of C20 or longer acyl chains.
Molluscs
Evidence confirms that molluscs possess elongases involved in the biosynthetic pathways of PUFA including NMI FA in bivalves such as Mesodesma mactroides (De Moreno et al. 1976), Crassostrea gigas (Waldock and Holland 1984), Megangulus zyonoensis, Scapharca (Anadara) broughtonii, Callista (Ezocallista) brevisiphonata and Mytilus edulis (Zhukova 1986, 1991; Kawashima and Ohnishi 2004), as well as gastropods like Haliotis fulgens (Durazo-Beltran et al. 2003). The above studies typically involved enzymatic activity assays using radiolabelled substrates or determination of FA composition of individuals subjected to controlled experimental conditions. However, further molecular evidence targeting the specific gene responsible for PUFA elongation has become available for a variety of mollusc species. Thus, a cDNA encoding an Elovl protein was first isolated from O. vulgaris (Monroig et al. 2012b). This protein was termed “Elovl5/2” or “Elovl2/5” (Monroig et al. 2013b, 2016b; Liu et al. 2013) based on its phylogenetic relationship with the vertebrate Elovl2 and Elovl5 elongases. Similar proteins were subsequently characterised from S. officinalis (Monroig et al. 2016a), C. nobilis (Liu et al. 2013) and Crassostrea angulata (Zhang et al. 2018). Regarding its function, the mollusc Elovl2/5 can efficiently elongate C18–20 PUFA substrates, but has no activity towards C22 substrates. Thus, despite the mollusc Elovl2/5 representing an ancestral protein of vertebrate Elovl5 and Elovl2 (Monroig et al. 2016b), this enzyme shows substrate specificities that resemble those of Elovl5 but not Elovl2. Indeed, Elovl2 is regarded as a key enzyme in the biosynthesis of DHA via the Sprecher pathway, since it can catalyse the elongation of 22:5n-3 to 24:5n-3, before subsequent ∆6 desaturation and partial β-oxidation lead to the production of DHA (Fig. 1) (Sprecher 2000). Consequently, lack of C22 elongation capacity within the mollusc Elovl2/5 has been hypothesised as one of the reasons accounting for the inability of cephalopods to biosynthesise DHA (Fig. 1) (Monroig et al. 2012b, 2013b). To clarify whether alternative Elovl enzymes could compensate for lack of ability to elongate 22:5n-3 to 24:5n-3 by Elovl2/5, an Elovl4-like cDNA was cloned and functionally characterised from O. vulgaris (Monroig et al. 2017). In agreement with functions of fish orthologues (Jin et al. 2017; Oboh et al. 2017b), the common octopus Elovl4 showed the ability to elongate 22:5n-3 to 24:5n-3. In spite of the elongase capacity of Elovl4, DHA biosynthesis appears not to be possible in octopus since, as described above, key desaturation activities such as ∆6 or ∆4 required for the two DHA biosynthetic pathways known in animals appear to be absent (Monroig et al. 2012a, 2017).
The octopus Elovl4 was able to produce very long-chain (> C24) PUFA that reached, in some cases, up to C34 (e.g., 34:5n-3; Monroig et al. 2017). The elongation abilities of the octopus Elovl4 were largely in agreement with those reported in teleost fish Elovl4 (Castro et al. 2016; Jin et al. 2017; Oboh et al. 2017b), but in disagreement with functional characterisation data from the C. nobilis Elovl4, which did not produced PUFA beyond C24. While the reasons for such an apparent discrepancy between functions of Elovl4 from O. vulgaris and C. nobilis remain unclear, it is worth noting that these existed despite both studies having used a very similar expression system based on yeast S. cerevisiae, arguably the most commonly used system to test the activity of enzymes involved in the biosynthesis of PUFA from a range of organisms including mammals (Park et al. 2009), fish (Hastings et al. 2001), plants (Pirtle et al. 2001), fungi (Sakuradani et al. 1999) and invertebrates (Peyou-Ndi et al. 2000). Generally, the yeast assay shows results of enzyme activity that match well those obtained by enzyme activity assays using labelled FA substrates. For instance, the elongase activities from cephalopod Elovl2/5 and Elovl4 described above are consistent with radiolabelled elongation products recovered from hatchlings of O. vulgaris (Reis et al. 2014) and S. officinalis (Reis et al. 2016) incubated with [1 − 14C]-labelled substrates. Studies in mammals have shown that very long-chain PUFA accumulate in relatively small amounts and specific lipid classes of certain tissues, and play important functions in vision and reproduction (Agbaga et al. 2010; McMahon and Kedzierski 2010; Zadravec et al. 2011). Roles of Elovl4 from invertebrates are not yet fully understood, but tissue distribution data showing high expression of the O. vulgaris Elovl4 in eye and gonads suggested these are also major sites for very long-chain PUFA biosynthesis in cephalopod molluscs (Monroig et al. 2017). Furthermore, it is also reasonable to expect that invertebrate Elovl4 is involved in the biosynthesis of very long-chain NMI FA typically found in marine invertebrates (Barnathan 2009; Kornprobst and Barnathan 2010).
Crustaceans
The ability of certain crustaceans for PUFA elongation has been evidenced through different methodological approaches. Indirect evidence based on FA analysis of both animals and diets strongly suggested that some marine copepods (Desvilettes et al. 1997; Nanton and Castell 1998; Parrish et al. 2012) and lobsters such as Macrobrachium rosenbergii, specifically its postlarval stages, were able to elongate PUFA (Reigh and Stickney 1989). Using liposomes formulated with D5-18:3n-3 for marine zooplankton species including C. finmarchicus, C. acutus, D. forcipatus and larvae of E. superba resulted in neglectable levels of metabolic PUFA products, with 20:3n-3 arising as the sole FA produced at a relatively higher rate (Bell et al. 2007). While the contribution of bacteria towards such elongation products could not be determined, it is now known that crustaceans possess Elovl genes that may account for some of the elongation activities reported in the studies above. Recently, an Elovl4 from the mud crab S. paramamosain has been investigated (Lin et al. 2018). The S. paramamosain Elovl4, isolated from hepatopancreas, encodes a protein of 338 amino acids with all common features of Elovl protein family members (Jakobsson et al. 2006). The phylogenetic analyses revealed that Elovl4 is found in other arthropods including aquatic crustaceans such as Daphnia magna and Hyalella azteca. Despite no functional data being presented, this elongase was upregulated with increased inclusion of vegetable oil in the diet, suggesting putative roles in PUFA biosynthesis since it exhibited similar regulatory mechanisms as homologous enzymes from aquatic vertebrates (Li et al. 2017). Also in agreement with vertebrate Elovl4, the mud crab S. paramamosain Elovl4 was highly expressed in cranial ganglia and the eyestalk, tissues in which typical products of elongation by Elovl4, namely very long-chain PUFA, accumulate and play important roles in neural function and vision. The presence of alternative elongases such as Elovl2/5 homologues is likely to occur in some crustaceans since elongation capacity towards C18–20 PUFA substrates was demonstrated in the copepod P. parvus (Moreno et al. 1979) and elongases homologous to vertebrate Elovl2 (gb|ACO10776.1|) can be identified in the genome of ectoparasitic copepod Caligus rogercresseyi (Monroig et al. 2013b).
Other invertebrates
No gene-encoding PUFA elongases have been functionally characterised in polychaetes, although Monroig et al. (2013b) retrieved in silico putative Elovl2- and Elovl5-like elongase sequences from C. teleta. Given the relative abundance of EPA in species such as Hediste (formerly known as Nereis) diversicolor, Perinereis nuntia, Laetmonice sp. and Paradiopatra sp., and the very low DHA content (Pairohakul 2013), it can be speculated that polychaetes possess PUFA elongases with low affinity towards C20 or longer PUFA substrates. With regards to echinoderms, a gene annotated as “Elovl5” has been cloned and functionally characterised from the sea cucumber A. japonicus (Li et al. 2016). While the deduced amino acid sequence contains all characteristic elements of the Elovl protein family, its categorisation as Elovl5 is not entirely clear from the phylogenetic analysis (Li et al. 2016). Thus, the A. japonicus Elovl5 did not cluster together with Elovl2/5 from molluscs and amphioxus, as it would have been expected. Irrespective of its phylogeny, the elongase characterised from A. japonicus appears to have a role in PUFA metabolism since it showed the ability to elongate γ-linolenic acid (18:3n-6) and EPA to their corresponding + 2C products (Li et al. 2016). It is interesting to note that the functional characterisation assays were run using Pichia pastoris, the same heterologous system used for the desaturase characterised from the same species and discussed above (Li et al. 2016; Liu et al. 2017b).
Invertebrate chordates, comprising amphioxus and tunicates, have been well-consolidated models in evolutionary developmental biology research and good quality genomes have existed for years. However, our knowledge of PUFA biosynthesis remains rather limited, particularly for front-end desaturases. With regards to elongases, an Elovl with roles in PUFA biosynthesis was characterised from the tunicate Ciona intestinalis (Meyer et al. 2004). This gene is a paralogue of Elovl4 as phylogenetic analyses established that this protein grouped together with the human and mouse ELOVL4 and separately from vertebrate Elovl2 and Elovl5 (Meyer et al. 2004). Its functional characterisation in yeast S. cerevisiae revealed the C. intestinalis Elovl4 was capable of elongating a range of PUFA substrates with chain lengths from C18 to C22, and with both 18:4n-3 and EPA as the most preferred substrates (Meyer et al. 2004). More recently, an elongase Elovl2/5 orthologous to that of molluscs was characterised from the European amphioxus Branchiostoma lanceolatum (Monroig et al. 2016b). Like the mollusc Elovl2/5, the amphioxus orthologue efficiently elongates C18 and C20 PUFA but, in addition, had some elongase activity towards C22 substrates (Monroig et al. 2016b). It is interesting to note that no orthologues of Elovl2/5 were found in genomes from sea squirts (C. intestinatis and Ciona savignyi) and the star ascidian (Botryllus schlosseri; Monroig et al. 2016b), thus leaving the abovementioned Elovl4 (Meyer et al. 2004) as the sole Elovl with expected roles in tunicate PUFA biosynthesis.
Regulation of PUFA-biosynthesising desaturases and elongases
Aquatic invertebrates have shown the ability to modulate their PUFA biosynthetic capacity through their diet (nutritional regulation) and environmental factors including temperature, pressure and salinity. Nutritional regulation of the PUFA biosynthesis pathways has been investigated in a variety of farmed species in the context of development of sustainable feed formulations with reduced inclusion of the finite marine ingredients fishmeal and fish oil, and increased levels of alternative ingredients such as vegetable oil. Generally, increased dietary levels of vegetable oil (devoid of ≥ C20 PUFA) results in an upregulation of both desaturase and Elovl genes. It has been hypothesised that such regulatory mechanism can, to some extent, compensate for the lower dietary level of ≥ C20 PUFA in vegetable oil-based diets. However, the specific molecular mechanisms involved in the regulation of desaturase and Elovl expression is poorly understood in invertebrates. A desaturase (termed “∆6”) and an elongase (termed “elongase-2”) from the Jade Tiger hybrid abalone (Haliotis rubra x H. laevigata) were upregulated with increasing levels of flaxseed oil (Mateos et al. 2012a) and canola oil (Mateos et al. 2012b). Despite such regulatory pattern being consistent with those observed in vertebrates (Monroig et al. 2018), it is important to note that neither the desaturase nor the elongase from the Jade Tiger hybrid were functionally characterised and, therefore, their role in PUFA biosynthesis cannot be fully established. Additionally, nomenclature of the genes in the abovementioned studies on the Jade Tiger hybrid abalone was not always consistent with that of vertebrate genes, this probably illustrating the complex phylogeny of the desaturase and elongase lineages existing in invertebrates. Such phylogenetic complexity often leads to misleading nomenclature of desaturases and elongases. For instance, a recent study investigating the responses of Macrobrachium nipponense-fed diets with increasing levels of ALA (Luo et al. 2018) considered a sphingolipid ∆4 desaturases as a front-end ∆4 desaturases. In the same study, the authors implied that Elovl6 might have a role in PUFA elongation on the basis that this gene was upregulated in individuals fed ALA-rich diets (Luo et al. 2018). Interestingly, an increased expression in response to dietary ALA was also observed for a M. nipponense desaturase termed as “∆6 fatty acyl desaturase” (Luo et al. 2018). This gene appears to be a paralogue of those mentioned above from other crustaceans, namely E. sinensis (Yang et al. 2013), S. paramamosain (Lin et al. 2017), L. vannamei (Chen et al. 2017) and C. quadricarinatu (Wu et al. 2018). Like observed in M. nipponense (Luo et al. 2018), the desaturases from the crabs E. sinensis (Yang et al. 2013), Portunus trituberculatus (Wang et al. 2014) and S. paramamosain (Lin et al. 2017), as well as that from spiny lobster Sagmariasus verreauxi (Shu-Chien et al. 2017) were also nutritionally upregulated by dietary vegetable oil, suggesting putative roles in PUFA metabolism.
Invertebrates are poikilotherms and their body temperature reflects that of the environment. Consequently, it is expected that exposure of aquatic invertebrates to low temperature results in changes of phospholipid FA composition as a mechanism aiming at maintaining membrane fluidity. Such changes in FA composition under low-temperature conditions have been reported in molluscs (Hall et al. 2002) and crustaceans (Pond 2012; Pond et al. 2014; Schlechtriem et al. 2006). Within the latter, the responses of the cyclopoid copepod Paracyclopina nana to low temperatures were investigated at a molecular level, including expression analyses of lipogenic genes including several desaturases and elongases (Lee et al. 2017a). While gene nomenclature used does not allow us to identify the specific enzyme, three fatty acyl elongases (“Elongase1–3”) were upregulated at temperature below ambient (20 °C) and downregulated under temperatures above. With regards to desaturases, the so-called “∆5-desaturase” and “∆9-desaturase” responded in a similar manner as elongases but the “∆4-desaturase” did not, probably explained by the fact that this enzyme is a sphingolipid ∆4 desaturase homologue. The activtion of these FA-biosynthesising genes generally correlated well with abundance of the enzymatic products, i.e., PUFA, in the lipids of P. nana (Lee et al. 2017a). Such alteration of the lipid composition, known as “homeoviscous adaptation” (Guschina and Harwood 2006b), occurs not only in response to lower temperature but also pressure (Pond et al. 2014). Indeed, a comparison of two closely related species of marine nematodes, namely Halomonhystera disjuncta occupying intertidal habitats and H. hermesi from deep-sea habitats, revealed that the FA elongation pathway was more prominent in H. hermesi, suggesting that this species adapts to high pressure by biosynthesising PUFA (Van Campenhout and Vanreusel 2016).
Salinity has been also described as an environmental factor that influences the biosynthesis of PUFA in aquatic organisms like fish (Vagner and Santigosa 2011). Despite the mechanism not being fully understood, exposure of fish to different salinities is believed to trigger an osmoregulatory response required for adaptation that is associated with membrane lipid remodelling to ensure normal function of membrane-bound proteins (Fonseca-Madrigal et al. 2012). However, whereas species like the pike silverside Chirostorma estor reduced the enzymatic activities of PUFA biosynthesis at lower salinities (Fonseca-Madrigal et al. 2012), other species like Siganus canaliculatus enhanced the PUFA biosynthesising genes when subjected to reduced salinity (Xie et al. 2015). The latter response appears to operate in the monogonont rotifer Brachionus koreanus, in which elongases and desaturases, some of which have putative roles in PUFA biosynthesis, were downregulated after exposure to high salinity (Lee et al. 2017b).
Concluding remarks
Data combining analytical, biochemical and molecular evidence have demonstrated that invertebrates can produce PUFA endogenously. At a molecular level, three major types of enzymes, namely ωx desaturases, front-end desaturases and Elovl enzymes, have been isolated and, in many cases, functionally characterised from sponges, cnidarians, molluscs, annelids, crustaceans, rotifers, echinoderms and non-vertebrate chordates. Front-end desaturase genes have been cloned and functionally characterised from molluscs and echinoderms, with substrate specificities including Δ5 and, less commonly, Δ6 and Δ8. Coexistence of Δ8/Δ5 and Δ8/Δ6 desaturase activities reported in C. nobilis and S. constricta, respectively, enable the EPA and ARA biosynthesis in some molluscs. However, such a metabolic ability appears to be missing in other species with one sole front-end desaturase (e.g., Octopus). Similarly, desaturases enabling Δ8 and Δ5 activities coexist in the sea urchin P. lividus. Phylogenetic analyses have not allowed us to understand yet whether these invertebrate front-end desaturases are orthologues of the vertebrate front-end desaturase, i.e., Fads (Lopes-Marques et al. 2018). More clearly though, the presence of invertebrate-specific front-end desaturase families are widespread among several phyla. This is the case of FadsA, consistently showing Δ5 desaturase activity, and confirmed to be present in molluscs, annelids, echinoderms and hemichordates (Surm et al. 2015; Kabeya et al. 2017b). Other invertebrate front-end desaturases appear to have a more restricted distribution and can be uniquely found in specific groups. An example of this is represented by FadsC, a group of front-end desaturases that can be found exclusively in sea urchins (Kabeya et al. 2017b). Importantly, these results emphasise the diversity of animal front-end desaturases.
Elovl-like enzymes with roles in PUFA biosynthesis have been also characterised in a variety of invertebrates. The previously characterised mollusc Elovl2/5 elongase (Monroig et al. 2012b) has been described in other invertebrate groups such as amphioxus. While the amphioxus Elovl2/5 showed some elongase activity towards C22 PUFA substrates, more typically invertebrate Elovl2/5 have C18 and C20 PUFA as preferred substrates for elongation (Monroig et al. 2012b, 2016a, b). Since biosynthesis of DHA often involves production of 24:5n-3 (Sprecher 2000), it is interesting to investigate alternative Elovl-like families with the ability to elongate C22 has been prompted. The mollusc Elovl4 has been demonstrated to elongate 22:5n-3 to 24:5n-3 and thus compensates for the lack of C22 elongase activity within Elovl2/5 (Liu et al. 2013; Monroig et al. 2017). The tunicate C. intestinalis has an Elovl4, but appears to lack other PUFA elongases such as Elovl2/5. While still fragmented information, our current knowledge of the PUFA elongase repertoire in invertebrates suggests that, while the presence of Elovl2/5 is more restricted, Elovl4 has a more widespread distribution and appears as a distinct gene in cnidarians (Surm et al. 2018), molluscs (Monroig et al. 2017) and crustaceans (Lin et al. 2018). Interestingly, the phylogenetic analysis appears to delineate the A. queenslandica protein Elovl1/2/4/5/7 as a basal sequence of the Elovl2/5 and Elovl4 found in more recently emerged invertebrate groups. As mentioned above for front-end desaturases, invertebrate-specific Elovl-like families might also exist as suggested by Surm et al. (2018) in Cnidaria.
Arguably, one of the most important discoveries in this research field has been the demonstration that methyl-end or ωx desaturases are widespread among invertebrates (Kabeya et al. 2018a). These results challenged the historically accepted dogma establishing that global production of n-3 PUFA occurred almost exclusively by marine microbes since it demonstrated that multiple invertebrates possess ωx desaturases enabling the biosynthesis of n-3 PUFA de novo and, from them, long-chain PUFA. Beyond the obvious ecological implications associated with primary production of essential nutrients at a global scale, the discovery of animal ωx desaturases has evidenced the need to revise extensively the PUFA biosynthetic pathways in invertebrates since, clearly, these can differ remarkably from vertebrate pathways. Often the invertebrate PUFA biosynthetic pathways as a whole, or the functions of specific enzymes in particular, have been inferred from studies on vertebrate species. From this review, it appears clear that such an approach might often lead to wrong interpretations given the complex phylogeny and functional diversification of desaturases and elongases found in invertebrates, along with the presence of family genes that are exclusive of certain invertebrate groups and whose functions are yet to be determined. In some instances, automatic gene annotation has led to misleading identification of candidate desaturases and elongases with non-demonstrated roles in PUFA biosynthesis, but often with regulatory responses that resemble those of well-established PUFA-biosynthesising genes. This highlights the need to run appropriate phylogenetic studies to allow the confident reconstruction of the ancestral state of each desaturase/elongase gene at key nodes in the animal phylogeny, which in turn permits discovery of molecular innovations in distinct major groups of animals. Equally important, functional characterisation of the desaturase and elongase genes becomes an essential tool to confirm which specific reactions the candidate enzymes carry out within the PUFA biosynthetic pathways.
References
Abdul Hamid NK, Carmona-Antonanzas G, Monroig Ó, Tocher DR, Turchini GM, Donald JA (2016) Isolation and functional characterisation of a fads2 in rainbow trout (Oncorhynchus mykiss) with Δ5 desaturase activity. PLoS One 11:e0150770
Agbaga MP, Mandal MNA, Anderson RE (2010) Retinal very long-chain PUFAs: new insights from studies on ELOVL4 protein. J Lipid Res 51:1624–1642
Ahlgren G, Vrede T, Goedkoop W (2009) Fatty acid ratios in freshwater fish, zooplankton and zoobenthos—are there specific optima? In: Arts MT et al (eds) Lipids in aquatic ecosystems. Springer, New York, pp 147–178
Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, Rokhsar DS (2015) The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524:220–224
Barclay WR, Meager KM, Abril JR (1994) Heterotrophic production of long chain omega-3 fatty acids utilizing algae and algae-like microorganisms. J Appl Phycol 6:123–129
Barnathan G (2009) Non-methylene-interrupted fatty acids from marine invertebrates: occurrence, characterization and biological properties. Biochimie 91:671–678
Bell MV, Dick JR, Anderson TR, Pond DW (2007) Application of liposome and stable isotope tracer techniques to study polyunsaturated fatty acid biosynthesis in marine zooplankton. J Plankton Res 29:417–422
Brett MT, Müller-Navarra DC, Persson J (2009) Crustacean zooplankton fatty acid composition. In: Arts MT et al (eds) Lipids in aquatic ecosystems. Springer, New York, pp 115–146
Brouwer IA, Geelen A, Katan MB (2006) n-3 Fatty acids, cardiac arrhythmia and fatal coronary heart disease. Prog Lipid Res 45:357–367
Brüggemann J (2012) Nematodes as live food in larviculture—a review. J World Aquacult Soc 43:739–763
Buckner JS, Hagen MM (2003) Triacylglycerol and phospholipid fatty acids of the silverleaf whitefly: composition and biosynthesis. Arch Insect Biochem Physiol 53:66–79
Carballeira NM, Maldonado ME (1989) On the isolation of the new fatty acid 6,11-eicosadienoic (20∶2) and related 6,11-dienoic acids from the sponge Euryspongia rosea. Lipids 24:665–668
Castro LFC, Monroig Ó, Leaver MJ, Wilson J, Cunha I, Tocher DR (2012) Functional desaturase Fads1 (Δ5) and Fads2 (Δ6) orthologues evolved before the origin of jawed vertebrates. PLoS ONE 7:e31950
Castro LFC, Tocher DR, Monroig Ó (2016) Long-chain polyunsaturated fatty acid biosynthesis in chordates: insights into the evolution of Fads and Elovl gene repertoire. Prog Lipid Res 62:25–40
Chapman JA et al (2010) The dynamic genome of Hydra. Nature 464:592–596
Chen K, Li E, Li T, Xu C, Xu Z, Qin JG, Chen L (2017) The expression of the Δ6 fatty acyl desaturase-like gene from Pacific white shrimp (Litopenaeus vannamei) under different salinities and dietary lipid compositions. J Shellfish Res 36:501–509
De Moreno JEA, Moreno VJ, Brenner RR (1976) Lipid metabolism of the yellow clam, Mesodesma mactroides: 2-polyunsaturated fatty acid metabolism. Lipids 11:561–566
De Troch M, Boeckx P, Cnudde C, Van Gansbeke D, Vanreusel A, Vincx M, Caramujo MJ (2012) Bioconversion of fatty acids at the basis of marine food webs: insights from a compound-specific stable isotope analysis. Mar Ecol Prog Ser 465:53–67
Desvilettes C, Bec A (2009) Formation and transfer of fatty acids in aquatic microbial food webs: role of heterotrophic protists. In: Arts MT et al (eds) Lipids in aquatic ecosystems. Springer, New York, pp 25–42
Desvilettes C, Bourdier G, Breton JC (1997) On the occurrence of a possible bioconversion of linolenic acid into docosahexaenoic acid by the copepod Eucyclops serrulatus fed on microalgae. J Plankton Res 19:273–278
Díaz-Almeyda E, Thomé PE, El Hafidi M, Iglesias-Prieto R (2011) Differential stability of photosynthetic membranes and fatty acid composition at elevated temperature in Symbiodinium. Coral Reefs 30:217–225
Durazo-Beltran E, D’Abramo LR, Toro-Vazquez JF, Vasquez-Pelaez C, Viana MT (2003) Effect of triacylglycerols in formulated diets on growth and fatty acid composition in tissue of green abalone (Haliotis fulgens). Aquaculture 224:257–270
Farkas T, Kariko K, Csengeri I (1981) Incorporation of [1 − 14C] acetate into fatty acids of the crustaceans Daphnia magna and Cyclops strenus in relation to temperature. Lipids 16:418–422
Ferraz RB, Kabeya N, Lopes-Marques M, Machado AM, Ribeiro RA, Salaro AL, Ozório R, Castro LFC, Monroig Ó (2018) A complete enzymatic capacity for long-chain polyunsaturated fatty acid biosynthesis is present in the Amazonian teleost tambaqui. Comp Biochem Physiol B (in press), Colossoma macropomum
Fonseca-Madrigal J, Pineda-Delgado D, Martínez-Palacios C, Rodríguez C, Tocher DR (2012) Effect of salinity on the biosynthesis of n-3 long-chain polyunsaturated fatty acids in silverside Chirostoma estor. Fish Physiol Biochem 38:1047–1057
Fonseca-Madrigal J, Navarro JC, Hontoria F, Tocher DR, Martinez-Palacios CA, Monroig Ó (2014) Diversification of substrate specificities in teleostei Fads2: characterization of Δ4 and Δ6Δ5 desaturases of Chirostoma estor. J Lipid Res 55:1408–1419
Gold DA, O’Reilly SS, Watson J, Degnan BM, Degnan SM, Krömer JO, Summons RE (2017) Lipidomics of the sea sponge Amphimedon queenslandica and implication for biomarker geochemistry. Geobiology 15:836–843
González-Rovira A, Mourente G, Zheng X, Tocher DR, Pendón C (2009) Molecular and functional characterization and expression analysis of a Δ6 fatty acyl desaturase cDNA of European sea bass (Dicentrarchus labrax L.). Aquaculture 298:90–100
Guillou H, Zadravec D, Martin PGP, Jacobsson A (2010) The key roles of elongases and desaturases in mammalian fatty acid metabolism: insights from transgenic mice. Prog Lipid Res 49:186–199
Guschina IA, Harwood JL (2006a) Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res 45:160–186
Guschina IA, Harwood JL (2006b) Mechanisms of temperature adaptation in poikilotherms. FEBS Lett 580:5477–5483
Hahn S, Stoilov IL, Ha TBT, Raederstroff D, Doss GA, Li H-T, Djerassi C (1988) Biosynthetic studies of marine lipids. 17. The course of chain elongation and desaturation in long-chain fatty acids of marine sponges. J Am Chem Soc 110:8117–8124
Hahn S, Lam WK, Wu I, Silva CJ, Djerassi C (1989) Unusual pattern of fatty acid biosynthesis. Evidence for C-19 desaturase activity in freshwater sponges. J Biol Chem 264:21043–21046
Hall JM, Parrish CC, Thompson RJ (2002) Eicosapentaenoic acid regulates scallop (Placopecten magellanicus) membrane fluidity in response to cold. Biol Bull 202:201–203
Hamilton ML, Powers S, Napier JA, Sayanova O (2016) Heterotrophic production of omega-3 long-chain polyunsaturated fatty acids by trophically converted marine diatom Phaeodactylum tricornutum. Mar Drugs 14:E53
Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, Kanehisa M (2008) The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes. J Lipid Res 49:183–191
Hastings N, Agaba MK, Tocher DR, Leaver MJ, Dick JR, Sargent JR, Teale AJ (2001) A vertebrate fatty acid desaturase with Δ5 and Δ6 activities. Proc Natl Acad Sci USA 98:14304–14309
Imbs AB, Yakovleva IM, Latyshev NA, Pham LQ (2010) Biosynthesis of polyunsaturated fatty acids in zooxanthellae and polyps of corals. Russ J Mar Biol 36:452–457
Imbs AB, Yakovleva IM, Dautova TN, Bui LH, Jones P (2014) Diversity of fatty acid composition of symbiotic dinoflagellates in corals: evidence for the transfer of host PUFAs to the symbionts. Phytochemistry 101:76–82
Ito MK, Simpson KL (1996) The biosynthesis of ω3 fatty acids from 18:2ω6 in Artemia spp. Comp Biochem Physiol B 115:69–76
Jakobsson A, Westerberg R, Jacobsson A (2006) Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res 45:237–249
Jin M, Monroig Ó, Navarro JC, Tocher DR, Zhou Q (2017) Molecular and functional characterisation of two elovl4 elongases involved in the biosynthesis of very long-chain (> C24) polyunsaturated fatty acids in black seabream Acanthopagrus schlegelii. Comp Biochem Physiol B 212:41–50
Joseph JD (1982) Lipid composition of marine and estuarine invertebrates. Part II: Mollusca. Prog Lipid Res 21:109–153
Joseph JD (1989) Distribution and composition of lipids in marine invertebrates. In: Ackman RG (ed) Marine biogenic lipids, fats, and oils, vol 2. CRC Press Inc, Boca Raton, pp 49–143
Jøstensen J-P, Landfald B (1997) High prevalence of polyunsaturated-fatty-acid producing bacteria in arctic invertebrates. FEMS Microbiol Lett 151:95–101
Kabeya N, Yamamoto Y, Cummins SF, Elizur A, Yazawa R, Takeuchi Y, Haga Y, Satoh S, Yoshizaki G (2015) Polyunsaturated fatty acid metabolism in a marine teleost, Nibe croaker Nibea mitsukurii: functional characterization of Fads2 desaturase and Elovl5 and Elovl4 elongases. Comp Biochem Physiol B 188:37–45
Kabeya N, Yoshizaki G, Tocher DR, Monroig Ó (2017a) Diversification of Fads2 in finfish species: Implications for aquaculture. In: Cruz-Suárez LE et al (eds) Investigación y desarrollo en nutrición acuícola. Universidad Autónoma de Nuevo León, San Nicolás de los Garza, pp 338–362
Kabeya N, Sanz-Jorquera A, Carboni S, Davie A, Oboh A, Monroig Ó (2017b) Biosynthesis of polyunsaturated fatty acids in sea urchins: molecular and functional characterisation of three fatty acyl desaturases from Paracentrotus lividus (Lamark 1816). PLoS One 12:e0169374
Kabeya N, Fonseca MM, Ferrier DEK, Navarro JC, Bay LK, Francis DS, Tocher DR, Castro LFC, Monroig Ó (2018a) Genes for de novo biosynthesis of omega-3 polyunsaturated fatty acids are widespread in animals. Sci Adv 4:eaar6849
Kabeya N, Yevzelman S, Oboh A, Tocher DR, Monroig Ó (2018b) Essential fatty acid metabolism and requirements of the cleaner fish, ballan wrasse Labrus bergylta: defining pathways of long-chain polyunsaturated fatty acid biosynthesis. Aquaculture 488:199–206
Kanazawa A, Teshima S, Ono K (1979) Relationship between essential fatty acid requirements of aquatic animals and the capacity for bioconversion of linolenic acid to highly unsaturated fatty acids. Comp Biochem Physiol B 63:295–298
Kawashima H, Ohnishi M (2004) Identification of minor fatty acids and various nonmethylene-interrupted diene isomers in mantle, muscle, and viscera of the marine bivalve Megangulus zyonoensis. Lipids 39:265–271
Kayama M, Hirata M (1984) Fatty acid metabolism in blue crab, Portunus trituberculatus, with special reference to de novo synthesis and conversion. J Jpn Oil Chem Soc 33:426–434
Kornprobst JM, Barnathan G (2010) Demospongic acids revisited. Mar Drugs 8:2569–2577
Kuah MK, Jaya-Ram A, Shu-Chien AC (2016) A fatty acyl desaturase (fads2) with dual Δ6 and Δ5 activities from the freshwater carnivorous striped snakehead Channa striata. Comp Biochem Physiol A 201:146–155
Lee M-C, Park JC, Kim D-H, Kang S, Shin K-H, Park HG, Han J, Lee J-S (2017a) Interrelationship of salinity shift with oxidative stress and lipid metabolism in the monogonont rotifer Brachionus koreanus. Comp Biochem Physiol A 214:79–84
Lee S-H, Lee M-C, Puthumana J, Park JC, Kang S, Han J, Shin K-H, Park HG, Om AS, Lee J-S (2017b) Effects of temperature on growth and fatty acid synthesis in the cyclopoid copepod Paracyclopina nana. Fish Sci 83:725–734
Leonard AE, Pereira SL, Sprecher H, Huang Y-S (2004) Elongation of long-chain fatty acids. Prog Lipid Res 43:36–54
Li Y, Monroig Ó, Zhang L, Wang S, Zheng X, Dick JR, You C, Tocher DR (2010) Vertebrate fatty acyl desaturase with Δ4 activity. Proc Natl Acad Sci USA 107:16840–16845
Li M, Mai K, He G, Ai Q, Zhang W, Xu W, Wang J, Liufu Z, Zhang Y, Zhou H (2013) Characterization of two Δ5 fatty acyl desaturases in abalone (Haliotis discus hannai Ino). Aquaculture 416–417:48–56
Li W, Feng Z, Song X, Zhu W, Hu Y (2016) Cloning, expression and functional characterization of the polyunsaturated fatty acid elongase (ELOVL5) gene from sea cucumber (Apostichopus japonicus). Gene 593:217–224
Li S, Monroig O, Wang T, Yuan Y, Navarro JC, Hontoria F, Liao K, Tocher DR, Mai K, Xu W, Ai Q (2017) Functional characterization and differential nutritional regulation of putative Elovl5 and Elovl4 elongases in large yellow croaker (Larimichthys crocea). Sci Rep 7:2303
Lin Z, Hao M, Zhu D, Li S, Wen X (2017) Molecular cloning, mRNA expression and nutritional regulation of a Δ6 fatty acyl desaturase-like gene of mud crab, Scylla paramamosain. Comp Biochem Physiol B 208–209:29–37
Lin Z, Hao M, Huang Y, Zou W, Rong H, Wen X (2018) Cloning, tissue distribution and nutritional regulation of a fatty acyl Elovl4-like elongase in mud crab, Scylla paramamosain (Estampador, 1949). Comp Biochem Physiol B 217:70–78
Liu H, Zheng H, Wang S, Wang Y, Li S, Liu W, Zhang G (2013) Cloning and functional characterization of a polyunsaturated fatty acid elongase in a marine bivalve noble scallop Chlamys nobilis Reeve. Aquaculture 416:146–151
Liu H, Guo Z, Zheng H, Wang S, Wang Y, Liu W, Zhang G (2014a) Functional characterization of a Δ5-like fatty acyl desaturase and its expression during early embryogenesis in the noble scallop Chlamys nobilis Reeve. Mol Biol Rep 41:7437–7445
Liu H, Zhang H, Zheng H, Wang S, Guo Z, Zhang G (2014b) PUFA biosynthesis pathway in marine scallop Chlamys nobilis Reeve. J Agric Food Chem 62:12384–12391
Liu H, Wang H, Cai S, Zhang H (2017a) A novel ω3-desaturase in the deep sea giant tubeworm Riftia pachyptila. Mar Biotechnol (NY) 19:345–350
Liu X, Wang L, Feng Z, Song X, Zhu W (2017b) Molecular cloning and functional characterization of the fatty acid delta 6 desaturase (FAD6) gene in the sea cucumber Apostichopus japonicus. Aquacult Res 48:4991–5003
Liyana-Pathirana C, Shahidi F, Whittick A, Hooper R (2002) Lipid and lipid soluble components of gonads of green sea urchin (Strongylocentrotus droebachiensis). J Food Lipids 9:105–126
Lopes-Marques M, Kabeya N, Qian Y, Ruivo R, Santos M, Venkatesh B, Tocher DR, Castro LFC, Monroig Ó (2018) Retention of fatty acyl desaturase 1 (fads1) in Elopomorpha and Cyclostomata provides novel insights into the evolution of long-chain polyunsaturated fatty acid biosynthesis in vertebrates. BMC Evol Biol (in press)
Luo N, Ding ZL, Kong YQ, Zhang RF, Zhang YX, Wu CL, Jiang ZQ, Ye JY (2018) An evaluation of increasing linolenic acid level in the diet of Macrobrachium nipponense: lipid deposition, fatty acid composition and expression of lipid metabolism-related genes. Aquac Nutr 24:758–767
Lv F, Nie Q, Wang T, Wang A, Yang W, Liu F, Yu Y, Lv L (2016) The effects of five dietary lipid sources on growth, body composition and antioxidant parameters of the clamworm, Perinereis aibuhitensis. Aquacult Res 48:5472–5480
Malcicka M, Visser B, Ellers J (2017) An evolutionary perspective on linoleic acid synthesis in animals. Evol Biol 45:15–26
Mateos HT, Lewandowski PA, Su XQ (2012a) Effects of dietary fish oil replacement with flaxseed oil on tissue fatty acid composition and expression of desaturase and elongase genes. J Sci Food Agric 92:418–426
Mateos HT, Lewandowski PA, Su XQ (2012b) The effect of replacing dietary fish oil with canola oil on fatty acid composition and expression of desaturase and elongase genes in Jade Tiger hybrid abalone. Food Chem 131:1217–1222
McMahon A, Kedzierski W (2010) Polyunsaturated extremely long chain C28–C36 fatty acids and retinal physiology. Br J Ophthalmol 94:1127–1132
Meesapyodsuk D, Qiu X (2012) The front-end desaturase: structure, function, evolution and biotechnological use. Lipids 47:227–237
Meldal BHM, Debenham NJ, De Ley P, De Ley IT, Vanfleteren JR, Vierstraete AR, Bert W, Borgonie G, Moens T, Tyler PA, Austen MC, Blaxter ML, Rogers AD, Lambshead PJD (2007) An improved molecular phylogeny of the Nematoda with special emphasis on marine taxa. Mol Phylogenet Evol 42:622–636
Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J (2001) Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 293:290–293
Meyer A, Kirsch H, Domergue F, Abbadi A, Sperling P, Bauer J, Cirpus P, Zank TK, Moreau H, Roscoe TJ, Zähringer U, Heinz E (2004) Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J Lipid Res 45:1899–1909
Mohd-Yusof NY, Monroig Ó, Mohd-Adnan A, Wan KL, Tocher DR (2010) Investigation of highly unsaturated fatty acid metabolism in the Asian sea bass, Lates calcarifer. Fish Physiol Biochem 36:827–843
Monroig Ó, Rotllant J, Sanchez E, Cerda-Reverter JM, Tocher DR (2009) Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim Biophys Acta 1791:1093–1101
Monroig Ó, Zheng X, Morais S, Leaver MJ, Taggart JB, Tocher DR (2010a) Multiple genes for functional Δ6 fatty acyl desaturases (Fad) in Atlantic salmon (Salmo salar L.): gene and cDNA characterization, functional expression, tissue distribution and nutritional regulation. Biochim Biophys Acta 1801:1072–1081
Monroig Ó, Rotllant J, Cerda-Reverter JM, Dick JR, Figueras A, Tocher DR (2010b) Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim Biophys Acta 1801:1145–1154
Monroig Ó, Li Y, Tocher DR (2011a) Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp Biochem Physiol B 159:206–213
Monroig Ó, Navarro JC, Tocher DR (2011b) Long-chain polyunsaturated fatty acids in fish: recent advances on desaturases and elongases involved in their biosynthesis. In: Cruz-Suarez LE et al (eds) Proceedings of the XI international symposium on aquaculture nutrition. Universidad Autónoma de Nuevo León, Monterrey, pp 257–282
Monroig Ó, Navarro JC, Dick JR, Alemany F, Tocher DR (2012a) Identification of a Δ5-like fatty acyl desaturase from the cephalopod Octopus vulgaris (Cuvier 1797) involved in the biosynthesis of essential fatty acids. Mar Biotechnol 14:411–422
Monroig Ó, Guinot D, Hontoria F, Tocher DR, Navarro JC (2012b) Biosynthesis of essential fatty acids in Octopus vulgaris (Cuvier, 1797): molecular cloning, functional characterisation and tissue distribution of a fatty acyl elongase. Aquaculture 360–361:45–53
Monroig Ó, Tocher DR, Hontoria F, Navarro JC (2013a) Functional characterisation of a Fads2 fatty acyl desaturase with Δ6/Δ8 activity and an Elovl5 with C16, C18 and C20 elongase activity in the anadromous teleost meagre (Argyrosomus regius). Aquaculture 412–413:14–22
Monroig Ó, Tocher DR, Navarro JC (2013b) Biosynthesis of polyunsaturated fatty acids in marine invertebrates: recent advances in molecular mechanisms. Mar Drugs 11:3998–4018
Monroig Ó, Hontoria F, Varó I, Tocher DR, Navarro JC (2016a) Investigating the essential fatty acids in the common cuttlefish Sepia officinalis (Mollusca, Cephalopoda): molecular cloning and functional characterisation of fatty acyl desaturase and elongase. Aquaculture 450:38–47
Monroig Ó, Lopes-Marques M, Navarro JC, Hontoria F, Ruivo R, Santos MM, Venkatesh B, Tocher DR, Castro LFC (2016b) Evolutionary functional elaboration of the Elovl2/5 gene family in chordates. Sci Rep 6:20510
Monroig Ó, de Llanos R, Varó I, Hontoria F, Tocher DR, Puig S, Navarro JC (2017) Biosynthesis of polyunsaturated fatty acids in Octopus vulgaris: molecular cloning and functional characterisation of a stearoyl-CoA desaturase and an elongation of very long-chain fatty acid 4 protein. Mar Drugs 15:82
Monroig Ó, Tocher DR, Castro LFC (2018) Chapter 3—Polyunsaturated fatty acid biosynthesis and metabolism in fish. In: Burdge GC (ed) Polyunsaturated fatty acid metabolism. Academic Press and AOCS Press, London, pp 31–60
Morais S, Monroig Ó, Zheng X, Leaver MJ, Tocher DR (2009) Highly unsaturated fatty acid synthesis in Atlantic salmon: characterization of Elovl5- and Elovl2-like elongases. Mar Biotechnol 11:627–639
Morais S, Castanheira F, Martinez-Rubio L, Conceicao LE, Tocher DR (2012) Long chain polyunsaturated fatty acid synthesis in a marine vertebrate: ontogenetic and nutritional regulation of a fatty acyl desaturase with Δ4 activity. Biochim Biophys Acta 1821:660–671
Moreno VJ, De Moreno JEA, Brenner RR (1979) Fatty acid metabolism in the calanoid copepod Paracalanus parvus: 1. Polyunsaturated fatty acids. Lipids 14:313–317
Morris RJ, Sargent JR (1973) Studies on the lipid metabolism of some oceanic crustaceans. Mar Biol 22:77–83
Mortillaro JM, Pitt KA, Lee SY, Meziane T (2009) Light intensity influences the production and translocation of fatty acids by zooxanthellae in the jellyfish Cassiopea sp. J Exp Mar Biol Ecol 378:22–30
Mourente G (1996) In vitro metabolism of 14C-polyunsaturated fatty acids in midgut gland and ovary cells from Penaeus kerathurus Forskâl at the beginning of sexual maturation. Comp Biochem Physiol B 115:255–266
Muscatine L, Gates RD, LaFontaine I (1994) Do symbiotic dinoflagellates secrete lipid droplets? Limnol Oceanogr 39:925–929
Nanton DA, Castell JD (1998) The effects of dietary fatty acids on the fatty acid composition of the haractacoid copepod, Tisbe sp. for use as a live food for marine fish larvae. Aquaculture 163:251–261
Oboh A, Betancor MB, Tocher DR, Monroig Ó (2016) Biosynthesis of long-chain polyunsaturated fatty acids in the African catfish Clarias gariepinus: molecular cloning and functional characterisation of fatty acyl desaturase (fads2) and elongase (elovl2) cDNAs. Aquaculture 462:70–79
Oboh A, Kabeya N, Carmona-Antoñanzas G, Castro LFC, Dick JR, Tocher DR, Monroig Ó (2017a) Two alternative pathways for docosahexaenoic acid (DHA, 22:6n-3) biosynthesis are widespread among teleost fish. Sci Rep 7:3889
Oboh A, Navarro JC, Tocher DR, Monroig Ó (2017b) Elongation of very long-chain (> C24) fatty acids in Clarias gariepinus: cloning, functional characterization and tissue expression of elovl4 elongases. Lipids 52:837–848
Olive PJW, Duangchinda T, Ashforth E, Craig S, Ward A, Davies SJ (2009) Net gain of long-chain polyunsaturated fatty acids (PUFA) in a lugworm Arenicola marina bioturbated mesocosm. Mar Ecol Prog Ser 387:223–239
Pairohakul S (2013) Evidence for polyunsaturated fatty acid biosynthesis in the ragworm (Nereis virens) and the Lugworm (Arenicola marina). PhD dissertation. Newcastle University, Newcastle upon Tyne
Papina M, Meziane T, van Woesik R (2003) Symbiotic zooxanthellae provide the host-coral Montipora digitata with polyunsaturated fatty acids. Comp Biochem Physiol B 135:533–537
Park WJ, Kothapalli KSD, Lawrence P, Tyburczy C, Brenna JT (2009) An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Δ8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res 50:1195–1202
Parrish CC, French VM, Whiticar MJ (2012) Lipid class and fatty acid composition of copepods (Calanus finmarchicus, C. glacialis, Pseudocalanus sp., Tisbe furcata and Nitokra lacustris) fed various combinations of autotrophic and heterotrophic protists. J Plankton Res 34:356–375
Pereira SL, Leonard AE, Mukerji P (2003) Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids 68:97–106
Peyou-Ndi MM, Watts JL, Browse J (2000) Identification and characterization of an animal Δ12 fatty acid desaturase gene by heterologous expression in Saccharomyces cerevisiae. Arch Biochem Biophys 376:399–408
Pirini M, Manuzzi MP, Pagliarani A, Trombetti F, Borgatti AR, Ventrella V (2007) Changes in fatty acid composition of Mytilus galloprovincialis (Lmk) fed on microalgal and wheat germ diets. Comp Biochem Physiol B 147:616–626
Pirtle IL, Kongcharoensuntorn W, Nampaisansuk M, Knesek JE, Chapman KD, Pirtle RM (2001) Molecular cloning and functional expression of the gene for a cotton Δ-12 fatty acid desaturase (FAD2). Biochim Biophys Acta 1522:122–129
Pond DW (2012) The physical properties of lipids and their role in controlling the distribution of zooplankton in the oceans. J Plankton Res 34:443–453
Pond DW, Allen CE, Bell MV, Van Dover C, Fallick AE, Dixon DR, Sargent JR (2002) Origins of long-chain polyunsaturated fatty acids in the hydrothermal vent worms Ridgea piscesae and Protis hydrothermica. Mar Ecol Prog Ser 225:219–226
Pond DW, Tarling GA, Mayor DJ (2014) Hydrostatic pressure and temperature effects on the membranes of a seasonally migrating marine copepod. PLoS One 9:e111043
Qiu X, Hong H, MacKenzie SL (2001) Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexaenoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J Biol Chem 276:31561–31566
Ran Z, Xu J, Liao K, Li S, Chen S, Yan X (2018) Biosynthesis of polyunsaturated fatty acids in the razor clam Sinonovacula constricta: characterization of Δ5 and Δ6 fatty acid desaturases. J Agric Food Chem 66:4592–4601
Reigh RC, Stickney RR (1989) Effects of purified dietary fatty acids on the fatty acid composition of freshwater shrimp, Macrobrachium rosenbergii. Aquaculture 77:157–174
Reis DB, Acosta NG, Almansa E, Navarro JC, Tocher DR, Monroig Ó, Andrade JP, Sykes AV, Rodríguez C (2014) In vivo metabolism of unsaturated fatty acids in Octopus vulgaris hatchlings determined by incubation with 14C-labelled fatty acids added directly to seawater as protein complexes. Aquaculture 431:28–33
Reis DB, Rodríguez C, Acosta NG, Almansa E, Tocher DR, Andrade JP, Sykes AV (2016) In vivo metabolism of unsaturated fatty acids in Sepia officinalis hatchlings. Aquaculture 450:67–73
Russell NJ, Nichols DS (1999) Polyunsaturated fatty acids in marine bacteria–a dogma rewritten. Microbiology 145:767–779
Sakuradani E, Kobayashi M, Ashikari T, Shimizu S (1999) Identification of Δ12-fatty acid desaturase from arachidonic acid-producing Mortierella fungus by heterologous expression in the yeast Saccharomyces cerevisiae and the fungus Aspergillus oryzae. FEBS J 261:812–820
Schauer PS, Simpson KL (1985) Bioaccumulation and bioconversion of dietary labeled fatty acid in Artemia and winter flounder (Pseudopleurunectes americanus). Can J Fish Aquat Sci 42:1430–1438
Schlechtriem C, Tocher DR, Dick J, Becker K (2004) Incorporation and metabolism of fatty acids by desaturation and elongation in the nematode, Panagrellus redivivus. Nematology 6:783–795
Schlechtriem C, Arts MT, Zellmer ID (2006) Effect of temperature on the fatty acid composition and temporal trajectories of fatty acids in fasting Daphnia pulex (Crustacea, Cladocera). Lipids 41:397–400
Shu-Chien AC, Han W-Y, Carter GC, Fitzgibbon QP, Simon CJ, Kuah M-K, Battaglene SC, Codabaccus BM, Ventura T (2017) Effect of dietary lipid source on expression of lipid metabolism genes and tissue lipid profile in juvenile spiny lobster Sagmariasus verreauxi. Aquaculture 479:342–351
Sprecher H (2000) Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta 1486:219–231
Spychalla J, Kinney A, Browse J (1997) Identification of an animal ω-3 fatty acid desaturase by heterologous expression in Arabidopsis. Proc Natl Acad Sci USA 94:1142–1147
Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, Larroux C, Putnam NH, Stanke M, Adamska M, Darling A, Degnan SM, Oakley TH, Plachetzki DC, Zhai Y, Adamski M, Calcino A, Cummins SF, Goodstein DM, Harris C, Jackson DJ, Leys SP, Shu S, Woodcroft BJ, Vervoort M, Kosik KS, Manning G, Degnan BM, Rokhsar DS (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726
Surm JM, Prentis PJ, Pavasovic A (2015) Comparative analysis and distribution of omega-3 lcPUFA biosynthesis genes in marine molluscs. PLoS One 10:e0136301
Surm JM, Toledo TM, Prentis PJ, Pavasovic A (2018) Insights into the phylogenetic and molecular evolutionary histories of Fad and Elovl gene families in Actiniaria. Ecol Evol 8:5323–5335
Svetashev VI, Vysotskii MV (1998) Fatty acids of Heliopora coerulea and chemotaxonomic significance of tetracosapolyenoic acids in coelenterates. Comp Biochem Physiol B 119:73–75
Takagi T, Kaneniwa M, Itabashi Y, Ackman RG (1986) Fatty acids in echinoidea: unusualcis-5-olefinic acids as distinctive lipid components in sea urchins. Lipids 21:558–565
Tanomman S, Ketudat-Cairns M, Jangprai A, Boonanuntanasarn S (2013) Characterization of fatty acid delta-6 desaturase gene in Nile tilapia and heterogenous expression in Saccharomyces cerevisiae. Comp Biochem Physiol B 166:148–156
Tocher DR (2015) Omega-3 long-chain polyunsaturated fatty acids and aquaculture in perspective. Aquaculture 449:94–107
Turchini G, Ng W-K, Tocher DR (2010) Fish oil replacement and alternative lipid sources in aquaculture feeds. CRC Press, Boca Raton
Uttaro AD (2006) Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life 58:563–571
Vagner M, Santigosa E (2011) Characterization and modulation of gene expression and enzymatic activity of delta-6 desaturase in teleosts: a review. Aquaculture 315:131–143
Van Campenhout J, Vanreusel A (2016) Closely related intertidal and deep-sea Halomonhystera species have distinct fatty acid compositions. Helgol Mar Res 70:8
Van der Horst DJ (1973) Biosynthesis of saturated and unsaturated fatty acids in the pulmonate land snail Cepaea nemoralis (L.). Comp Biochem Physiol B 46:551–560
Van der Horst DJ (1974) In vivo biosynthesis of fatty acids in the pulmonate land snail Cepaea nemoralis (L.) under anoxic conditions. Comp Biochem Physiol B 47:181–187
Vonk JA, van Kuijk BF, van Beusekom M, Hunting ER, Kraak MHS (2016) The significance of linoleic acid in food sources for detritivorous benthic invertebrates. Sci Rep 6:35785
Waldock MJ, Holland DL (1984) Fatty acid metabolism in young oysters, Crassostrea gigas: polyunsaturated fatty acids. Lipids 19:332–336
Wang M, Chen H, Gu Z, Zhang H, Chen W, Chen YQ (2013) ω3 fatty acid desaturases from microorganisms: structure, function, evolution, and biotechnological use. Appl Microbiol Biotechnol 97:10255–10262
Wang W, Wu X, Liu Z, Zheng H, Cheng Y (2014) Insights into hepatopancreatic functions for nutrition metabolism and ovarian development in the crab Portunus trituberculatus: gene discovery in the comparative transcriptome of different hepatopancreas stages. PLoS One 9:e84921
Weinert J, Blomquist GJ, Borgeson CE (1993) De novo biosynthesis of linoleic acid in two non-insect invertebrates: the land slug and the garden snail. Experientia 49:919–921
Wu D-L, Huang Y-H, Liu Z-Q, Yu P, Gu P-H, Fan B, Zhao Y-L (2018) Molecular cloning, tissue expression and regulation of nutrition and temperature on Δ6 fatty acyl desaturase-like gene in the red claw crayfish (Cherax quadricarinatus). Comp Biochem Physiol B 225:58–66
Xie D, Wang S, You C, Chen F, Tocher DR, Li Y (2015) Characteristics of LC-PUFA biosynthesis in marine herbivorous teleost Siganus canaliculatus under different ambient salinities. Aquac Nutr 21:541–551
Xue Z, He H, Hollerbach D, Macool DJ, Yadav NS, Zhang H, Szostek B, Zhu Q (2013) Identification and characterization of new Δ-17 fatty acid desaturases. Appl Microbiol Biotechnol 97:1973–1985
Yang Z, Guo Z, Ji L, Zeng Q, Wang Y, Yang X, Cheng Y (2013) Cloning and tissue distribution of a fatty acyl Δ6-desaturase-like gene and effects of dietary lipid levels on its expression in the hepatopancreas of Chinese mitten crab (Eriocheir sinensis). Comp Biochem Physiol B 165:99–105
Zadravec D, Tvrdik P, Guillou H, Haslam R, Kobayashi T, Napier JA, Capecchi MR, Jacobsson A (2011) ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J Lipid Res 52:245–255
Zhang H, Liu H, Cheng D, Liu H, Zheng H (2018) Molecular cloning and functional characterisation of a polyunsaturated fatty acid elongase in a marine bivalve Crassostrea angulata. J Food Nutr Res 6:89–95
Zheng X, Seiliez I, Hastings N, Tocher DR, Panserat S, Dickson CA, Bergot P, Teale AJ (2004) Characterization and comparison of fatty acyl Δ6 desaturase cDNAs from freshwater and marine teleost fish species. Comp Biochem Physiol B 139:269–279
Zheng X, Ding Z, Xu Y, Monroig Ó, Morais S, Tocher DR (2009) Physiological roles of fatty acyl desaturases and elongases in marine fish: characterisation of cDNAs of fatty acyl Δ6 desaturase and elovl5 elongase of cobia (Rachycentron canadum). Aquaculture 290:122–131
Zhou X-R, Horne I, Damcevski K, Haritos V, Green A, Singh S (2008) Isolation and functional characterization of two independently-evolved fatty acid Δ12-desaturase genes from insects. Insect Mol Biol 17:667–676
Zhou X-R, Green AG, Singh SP (2011) Caenorhabditis elegans Δ12-Desaturase FAT-2 is a bifunctional desaturase able to desaturate a diverse range of fatty acid substrates at the Δ12 and Δ15 positions. J Biol Chem 286:43644–43650
Zhu N, Dai X, Lin DS, Connor WE (1994) The lipids of slugs and snails: evolution, diet and biosynthesis. Lipids 29:869–875
Zhukova NV (1986) Biosynthesis of non-methylene-interrupted dienoic fatty acids for [C-14] acetate in molluscs. Biochim Biophys Acta 878:131–133
Zhukova NV (1991) The pathway of the biosynthesis of non-methylene-interrupted dienoic fatty acids in mollusks. Comp Biochem Physiol B 100:801–804
Acknowledgements
N.K. was funded by the Japan Society for the Promotion of Science through a Grant-in-Aid for a JSPS Research Fellow.
Author information
Authors and Affiliations
Corresponding author
Additional information
JSPS KAKENHI Grant Number JP 262003.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Monroig, Ó., Kabeya, N. Desaturases and elongases involved in polyunsaturated fatty acid biosynthesis in aquatic invertebrates: a comprehensive review. Fish Sci 84, 911–928 (2018). https://doi.org/10.1007/s12562-018-1254-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12562-018-1254-x