Abstract
Five Oenococcus oeni strains, selected from spontaneous malolactic fermentation (MLF) of Patagonic Pinot noir wine, were assessed for their use as MLF starter cultures. After the individual evaluation of tolerance to some stress conditions, usually found in wine (pH, ethanol, SO2, and lysozyme), the behavior of the strains was analyzed in MLO broth with 14 % ethanol and pH 3.5 in order to test for the synergistic effect of high ethanol level and low pH and, finally, in a wine-like medium. Although the five strains were able to grow in MLO broth under low pH and/or high ethanol, they must be acclimated to grow in a wine-like medium. Additionally, glycosidase and tannase activities were evaluated, showing differences among the strains. The potential of the strains to ferment citrate was tested and two of the five strains showed the ability to metabolize this substrate. We did not detect the presence of genes encoding histidine, tyrosine descarboxylase, and putrescine carbamoyltransferase. All the strains tested exhibited good growth capacity and ability to consume l-malic acid in a wine-like medium after cell acclimation, and each of them showed a particular enzyme profile, which might confer different organoleptic properties to the wine.




Similar content being viewed by others
References
Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE (1992) DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res 20:5137–5142
Apostol BL, Black WC IV, Miller BR, Reiter P, Beaty BJ (1993) Estimation of the number of full sibling families at an oviposition site using RAPD–PCR markers: applications to the mosquito Aedes aegypti. Theor Appl Genetics 86:991–1000
Bae S, Fleet GH, Heards GM (2006) Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100:712–727
Bartowsky EJ (2005) Oenococcus oeni and malolactic fermentation—moving in the molecular arena. Aust J Grape Wine Res 11:174–187
Bartowsky EJ, Borneman AR (2011) Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine. Appl Microbiol Biotechnol 92:441–447
Beltramo C, Desroche N, Tourdot-Maréchal R, Grandvalet C, Guzzo J (2006) Real time PCR for characterizing the stress response of Oenococcus oeni in wine-like medium. Res Microbiol 157:267–274
Bokulich NA, Thorngate JH, Richardson PM, Mills DA (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci 111:139–148
Bravo-Ferrada BM, Delfederico L, Hollmann A, Valdés La Hens D, Curilén Y, Caballero A, Semorile L (2011) Oenococcus oeni from Patagonian red wines: isolation, characterization and technological properties. Int J Microbiol Res 3:48–55
Bravo-Ferrada BM, Hollmann A, Delfederico L, Valdés La Hens D, Caballero A, Semorile L (2013) Patagonian red wines: selection of Lactobacillus plantarum starter cultures for malolactic fermentation. World J Microbiol Biotechnol 29:1537–1549
Bravo-Ferrada BM, Tymczyszyn EE, Gómez-Zavaglia A, Semorile L (2014) Effect of acclimation medium on cell viability, membrane integrity and ability to consume malic acid in synthetic wine by oenological Lactobacillus plantarum strains. J Appl Microbiol 116:360–367
Bon E, Delaherche A, Bilhacre E, De Daruvar A, Lonvaud-Funel A, Le Marrec C (2009) Oenococcus oeni genome plasticity is associated with fitness. Appl Environ Microbiol 75:2079–2090
Bourdineaud JP (2006) Both arginine and fructose stimulate pH-independent resistance in the wine bacteria Oenococcus oeni. Int J Food Microbiol 107:274–280
Cafaro C, Bonomo MG, Rossano R, Larocca M, Salzano G (2014) Efficient recovery of whole cell proteins in Oenococcus oeni - a comparison of different extraction protocols for high throughput malolactic starter applications. Folia Microbiol 59:399–408
Carreté R, Reguant C, Rozés N, Constantí M, Bordons A (2006) Analysis of Oenococcus oeni strains in simulated microvinifications with some stress compounds. Am J Enol Vitic 57:356–362
Cecconi D, Milli A, Rinalducci S, Zolla L, Zapparoli G (2009) Proteomic analysis of Oenococcus oeni freeze-dried culture to assess the importance of cell acclimation to conduct malolactic fermentation in wine. Electrophoresis 30:2988–2995
Chu-Ky S, Tourdot-Marechal R, Marechal PA, Guzzo J (2005) Combined cold, acid, ethanol shocks in Oenococcus oeni: effects on membrane fluidity and cell viability. Biochim Biophys Acta 1717:118–124
Cocconcelli PS, Porro D, Galandini S, Senini L (1995) Development of RAPD protocol for typing of strains of lactic acid bacteria and enterococci. Appl Microbiol 21:376–379
Costantini A, Rantsiou K, Majumder A, Jacobsen S, Pessione E, Svensson B, Garcia-Moruno E, Cocolin L (2015) Complementing DIGE proteomics and DNA subarray analyses to shed light on Oenococcus oeni adaptation to ethanol in wine-simulated conditions. J Proteomics 18:114–127
da Silveira MG, San Romão V, Loureiro-Dias MC, Rombouts FM, Abee T (2002) Flow cytometry assesment of the membrane integrity of ethanol-stressed Oenococcus oeni cells. Appl Environ Microbiol 68:6087–6093
da Silveira MG, Golovina EA, Hoekstra FA, Rombouts FM, Abee T (2003) Membrane fluidity adjustments in ethanol-stressed Oenococcus oeni cells. Appl Environ Microbiol 69:5826–5832
da Silveira MG, Baumgartner M, Rombouts FM, Abee T (2004) Effect of adaptation to ethanol on cytoplasmic and membrane protein profiles of Oenococcus oeni. Appl Environ Microbiol 70:2748–2755
da Silveira MG, Abee T (2009) Activity of ethanol-stressed Oenococcus oeni cells: a flow cytometric approach. J Appl Microbiol 106:1690–1696
Davis CR, Wibowo D, Eschenbruch R, Lee TH, Fleet GH (1985) Practical implications of malolactic fermentation: a review. Am J Enol Vitic 36:290–301
Davis CR, Wibowo D, Fleet GH, Lee TH (1988) Growth and metabolism of lactic acid bacteria during and after malolactic fermentation of wines at different pH. Appl Environ Microbiol 51:539–545
Delfederico L, Hollmann A, Martínez M, Iglesias NG, De Antoni G, Semorile L (2006) Molecular identification and typing of lactobacilli isolated from kefir grains. J Dairy Res 73:20–27
Fleet GH, Heard GM (1993) Yeasts: growth during fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic, Chur, Switzerland, pp 27–54
Fugelsang KC, Edwards CG (1997) Wine microbiology: practical applications and procedures, 2nd edn. Springer, New York
G-Alegría E, López I, Ignacio Ruiz J, Sáenz J, Fernández E, Zarazaga M, Dizy M, Torres C, Ruiz-Larrea F (2004) High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett 230:53–61
Guerrini S, Bastiniani A, Blaiotta G, Granchi L, Moschetti G, Coppola S, Romano P, Vincenzini M (2003) Phenotypic and genotypic characterization of Oenococcus oeni strains isolated from typical Italian wines. Int J Food Microbiol 83:1–14
González-Arenzana L, Santamaría P, López R, Tenorio C, López-Alfaro I (2012a) Ecology of indigenous lactic acid bacteria along different winemaking processes of Tempranillo red wine from La Rioja (Spain). Scientific World J 2012:796327
Grimaldi A, McLean H, Jiranek V (2000) Identification and partial characterization of glycosidic activities of commercial strains of the lactic acid bacteria Oenococcus oeni. Am J Enol Vitic 51:362–369
Grimaldi A, Bartowsky E, Jiranek V (2005) Screening of Lactobacillus spp. and Pediococcus spp. for glycosidase activities that are important in oenology. J Appl Microbiol 99:1061–1069
González-Arenzana L, López R, Santamaría P, Tenorio C, López-Alfaro I (2012b) Dynamics of indigenous lactic acid bacteria populations in wine fermentations from la Rioja (Spain) during three vintages. Microb Ecol 63:12–19
Henick-Kling T (1995) Control of malolactic fermentation in wine: energetic, flavour modification and methods of starter culture preparation. J Appl Bacteriol Symp Suppl 79:29S–37S
Kempler GM, McKay LL (1980) Improved medium for detection of citrate-fermenting Streptococcus lactis subsp diacetylactis. Appl Environm Microbiol 39:926–927
Landete JM, Arena ME, Pardo I, Manca de Nadra MC, Ferrer S (2010) The role of two families of bacterial enzymes in putrescine synthesis from agmatine via agmatine deiminase. Int Microbiol 13:169–177
Le Jeune C, Lonvaud-Funel A, Ten Brink B, Hofstra H, van der Vossen JMBM (1995) Development of a detection system for histidine decarboxilating lactic acid bacteria based on DNA probes, PCR and activity test. Appl Environ Microbiol 78:316–326
Lechiancole T, Blaiotta G, Messina D, Fusco V, Villani F, Salzano G (2006) Evaluation of intra-specific diversities in Oenococcus oeni through analysis of genomic and expressed DNA. Syst Appl Microbiol 29:375–381
Lekha PK, Lonsane BK (1997) Production and application of tannin acyl hydrolase: state of the art. Adv Appl Microbiol 44:215–60
Lerm E, Engelbrecht L, du Toit M (2010) Malolactic fermentation: the ABC’s of MLF. S Afr J Enol Vitic 31:186–212
Lonvaud-Funel A (1999) Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 76:317–331
López R, López-Alfaro I, Gutiérrez AR, Tenorio C, Garijo P, González-Arenzana L, Santamaría P (2001) Malolactic fermentation of Tempranillo wines: contribution of the lactic acid bacteria to sensory quality and chemical composition. Int J Food Sci Technol 46:2373–2381
Lucas P, Landete J, Coton M, Coton E, Lonvaud-Funel A (2003) The tyrosine decarboxylase operon of Lactobacillus brevis IOEB 9809: characterization and conservation in tyramine-producing bacteria. FEMS Microbiol Lett 229:65–71
Lucas P, Lonvaud-Funel A (2002) Purification and parcial genes equence of the tyrosine decarboxylase of Lactobacillus brevis IOEB 9809. FEMS Microbiol Lett 211:85–89
Maicas S, González-Cabo P, Ferrer S, Pardo I (1999) Production of Oenococcus oeni biomass to induce malolactic fermentation in wine by control of pH and substrate addition. Biotechnol Lett 21:349–353
Martineau B, Henick-Kling T (1995) Formation and degradation of diacetyle in wine during alcoholic fermentation with Saccharomyces cerevisiae strain EC 1118 and malolactic fermentation with Leuconostoc œnos strain MCW. Am J Enol Vitic 46:442–448
Matthews A, Grimaldi A, Walker M, Bartowsky E, Grbin P, Jiranek V (2004) Lactic acid bacteria as a potential source of enzymes for use in vinification. Appl Environ Microbiol 70:5715–5731
Moreno-Arribas MV, Polo CM, Jorganes F, Muñoz R (2003) Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int J Food Microbiol 84:117–123
Osawa R, Walsh TP (1993) Visual reading method for detection of bacterial tannase. Appl Environ Microbiol 59:1251–1252
Renouf V, Claisse O, Lonvaud-Funel A (2007) Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol 75:149–164
Rodas AM, Ferrer S, Pardo I (2003) 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Syst Appl Microbiol 26:412–422
Rosi I, Fia G, Canuti V (2003) Influence of different pH values and inoculation time on the growth and malolactic activity of a strain of Oenococcus oeni. Aust J Grape Wine Res 9:194–199
Ruiz P, Seseña S, Izquierdo PM, Llanos Palop M (2010) Bacterial biodiversity and dynamics during malolactic fermentation of Tempranillo wines as determined by a culture-independent method (PCR-DGGE). Appl Microbiol Biotechnol 86:1555–1562
Sico MA, Bonomo MG, Salzano G (2008) Isolation and characterization of Oenococcus oeni from Aglianico wines. World J Microbiol Biotechnol 24:1829–1835
Solieri L, Giudici P (2010) Development of a sequence-characterizede amplified region marker-targeted quantitative PCR assay for strain-specific detection of Oenococcus oeni during wine malolactic fermentation. Appl Environ Microbiol 76:7765–7774
Solieri L, Genova F, De Paola M, Giudici P (2010) Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures. J Appl Microbiol 108:285–298
Stendid J, Karlsson JO, Hogberg N (1994) Intraspecific genetic variation in Heterobasidium mannosum revealed by amplification of minisatellite DNA. Mycol Res 98:57–63
Ugliano M, Genovese A, Moio L (2003) Hydrolysis of wine aroma precursors during malolactic fermentation with four commercial starter cultures of Oenococcus oeni. J Agric Food Chem 51:5073–5078
Ulrike E, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene encoding for 16S ribosomal RNA. Nucl Acids Res 17:7843–7853
Vaquero I, Marcobal A, Muñoz R (2004) Tannase activity by lactic acid bacteria isolated from grape must and wine. Int J Food Microbiol 96:199–204
Valdés La Hens D, Bravo-Ferrada B, Delfederico L, Caballero A, Semorile L (2015) Patagonian red wines: PCR-DGGE analysis with two targeted genes revealed the prevalence of Lactobacillus plantarum and Oenococcus oeni during spontaneous malolactic fermentation. Aust J Grape Wine R 21:49–56
Vigentini I, Picozzi C, Tirelli A, Giugni A, Foschino R (2009) Survey on indigenous Oenococcus oeni strains isolated from red wines of Valtellina, a cold climate wine-growing Italian area. Int J Food Microbiol 136:123–128
Yanagida F, Srionnual S, Chen YS (2008) Isolation and characteristics of lactic acid bacteria from koshu vineyards in Japan. Lett Appl Microbiol 47:134–139
Winterhalter P, Skouroumounis G (1997) Glycoconjugated aroma compounds: occurrence, role and biotechnological transformation. Adv Biochem Eng Biotechnol 55:73–105
Acknowledgments
This work was funded by grants from Universidad Nacional de Quilmes (Programa Microbiología Molecular Básica y Aplicaciones Biotecnológicas), Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CIC-PBA), and ANPCyT (PICTO UNQ 2006 N° 36474, PICT SU 2012 N° 2804, PICT 2013 N° 1481). BMBF and NB are fellows of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). LS is a member of the Research Career of CIC-BA; AH and ET are members of the Research Career of CONICET.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1: Supplementary 1
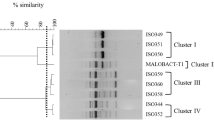
Clustering of O. oeni isolates from Patagonian Pinot noir wine. The dendrogram was obtained by UPGMA analysis of Coc, M13, and 1254 RAPD-PCR patterns. The reference strain is O. oeni ATCC 27310 = 27310. (JPG 1014 kb)
Rights and permissions
About this article
Cite this article
Bravo-Ferrada, B.M., Hollmann, A., Brizuela, N. et al. Growth and consumption of l-malic acid in wine-like medium by acclimated and non-acclimated cultures of Patagonian Oenococcus oeni strains. Folia Microbiol 61, 365–373 (2016). https://doi.org/10.1007/s12223-016-0446-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-016-0446-y