Abstract
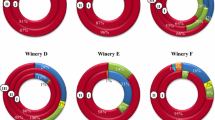
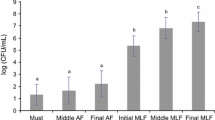
The bacterial population during malolactic fermentation of Tempranillo wine was studied using the polymerase chain reaction-denaturing gradient gel electrophoresis, a culture-independent method successfully used for identification and monitoring of bacterial population in different habitats included food fermentations. The results showed that Oenococcus oeni was the predominant species in the malolactic fermentation of Tempranillo wines, although the presence of Gluconobacter oxydans, Asaia siamensis, Serratia sp., and Enterobacter sp. was also observed. These results were partly coincidental with those obtained from a culture-dependent method, using a selective medium. Therefore, it may be concluded that for a more complete knowledge of the bacterial community present during malolactic fermentation of Tempranillo wine, an approach that combines a culture-independent method and a culture-dependent method would be advisable.

Similar content being viewed by others
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Ampe F, Sirvent A, Zakhia N (2001) Dynamics of the microbial community responsible for traditional sour cassava starch fermentation studied by denaturing gradient gel electrophoresis and quantitative rRNA hybridization. Int J Food Microbiol 65:45–54
Andorrà I, Landi S, Mas A, Guillamón JM, Esteve-Zarzoso B (2008) Effect of oenological practices on microbiol populations using culture-independent techniques. Food Microbiol 25(7):849–856
Bae S, Fleet GH, Heard GM (2006) Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100(4):712–725
Claus D, Lack P, Neu P (1983) D.S.M. catalogue of strains. Deutsche Sammlung yon Mikroorganismen
Coppola S, Fusco V, Andolfi R, Aponte M, Blaiotta G, Ercolini D, Moschetti G (2006) Evaluation of microbial diversity during the manufacture of Fior di Latte di Agerola, a traditional raw milk pasta-filata cheese of the Naples area. J Dairy Res 29:1–9
Endo A, Okada S (2005) Monitoring the lactic acid bacterial diversity during shochu fermentation by PCR-Denaturing gradient gel electrophoresis. J Biosci Bioeng 99:216–221
Ercolini D (2004) PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Methods 56:297–314
Giannino ML, Marzotto M, Dellaglio F, Feligini M (2009) Study of microbial diversity in raw milk and fresh curd used for Fontina cheese production by culture-independent methods. Int J Food Microbiol 130:188–195
Giraffa G, Neviani E (2001) DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int J Food Microbiol 67:19–34
Henick-Kling T (1993) Malolactic fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic, Switzerland, pp 289–326
Holt JH, Krieg NR, Sneath PHA, Staley JT, Williams ST (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins, Baltimore
Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180:4765–4774
Lechiancole T, Blaiotta G, Messina D, Fusco V, Villani F, Salzano G (2006) Evaluation of intra-specific diversities in Oenococcus oeni through analysis of genomic and expressed DNA. Syst Appl Microbiol 29:375–381
Li P, Wang Y, Wang Y, Liu K, Tong L (2010) Bacterial community structure and diversity during establishment of an anaerobic bioreactor to treat swine wastewater. Water Sci Technol 61(1):243–252
Liu FH, Lin GH, Gao G, Qin BQ, Zhang JS, Zhao GP, Zhou ZH, Shen JH (2009) Bacterial and archaeal assemblages in sediments of a large shallow freshwater lake, Lake Taihu, as revealed by denaturing gradient gel electrophoresis. J Appl Microbiol 106(3):1022–1032
Lonvaud-Funel A (1999) Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 76:317–331
Lopez I, Ruiz-Larrea F, Cocolin L, Orr E, Phister T, Marshall M, VanderGheynst J, Mills DA (2003) Design and evaluation of PCR primers for analysis of bacterial populations in wine by denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:6801–6807
Lubbs DC, Vester BM, Fastinger ND, Swanson KS (2009) Dietary protein concentration affects intestinal microbiota of adult cats: a study using DGGE and qPCR to evaluate differences in microbial populations in the feline gastrointestinal tract. J Anim Physiol Anim Nutr 93(1):113–121
Meroth CB, Walter J, Hertel C, Brandt MJ, Hammes WP (2003) Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol 69:475–482
Miambi E, Guyot JP, Ampe F (2003) Identification, isolation and quantification of representative bacteria from fermented cassava dough using an integrated approach of culture-dependent and culture-independent methods. Int J Food Microbiol 82:111–120
Ogier JC, Son O, Gruss A, Tailliez P, Delacroix-Buchet A (2002) Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl Environ Microbiol 68:3691–3701
Pérez Pulido R, Ben Omar N, Abriouel H, Lucas R, Martínez M, Gálvez A (2005) Microbiological study of lactic acid fermentation of caper berries by molecular and culture-dependent methods. Appl Environ Microbiol 71:7872–7879
Petersson A, Domig KJ, Nagel P, Zollitsch W, Hagmüller W, Kneifeld W (2009) Denaturing gradient gel electrophoresis (DGGE)-based monitoring of intestinal lactobacilli and bifidobacteria of pigs during a feeding trial. Arch Anim Nutr 63(2):112–126
Ponnusamy L, Xu N, Stav G, Wesson DM, Schal C, Apperson CS (2008) Diversity of bacterial communities in container habitats of mosquitoes. Microb Ecol 56(4):593–603
Prakitchaiwattana CJ, Fleet GH, Heard GM (2004) Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res 4:865–877
Puglisi E, Fragoulis G, Ricciuti P, Cappa F, Spaccini R, Piccolo A, Trevisan M, Crecchio C (2009) Effects of a humic acid and its size-fractions on the bacterial community of soil rhizosphere under maize (Zea mays L.). Chemosphere 77(6):829–837
Reguant C, Bordons A (2003) Typification of Oenococcus oeni strains by multiplex RAPD-PCR and study of population dynamics during malolactic fermentation. J Appl Microbiol 95(2):344–353
Renouf V, Claisse O, Lonvaud-Funel A (2005) Numeration, identification and understanding of microbial biofilm on grape berry surface. Aust J Grape Wine Res 11:316–327
Renouf V, Claisse O, Miot-Sertier C, Lonvaud-Funel A (2006) Lactic acid bacteria evolution during winemaking: use of rpoB gene as a target for PCR-DGGE analysis. Food Microbiol 23:136–145
Renouf V, Claisse O, Lonvaud-Funel A (2007) Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol 75:149–164
Rodas AM, Ferrer S, Pardo I (2005) Polyphasic study of wine Lactobacillus strains: taxonomic implications. Int J Syst Evol Microbiol 55:197–207
Ruiz P, Izquierdo PM, Seseña S, MLl P (2010) Analysis of lactic acid bacteria populations during spontaneous malolactic fermentation of Tempranillo wines at five wineries during two consecutive vintages. Food Control 21:70–75
Sánchez I, Seseña S, MLl P (2004) Polyphasic study of the genetic diversity of lactobacilli associated with “Almagro” eggplants spontaneous fermentation based on combined numerical analysis of randomly amplified polymorphic DNA and pulsed field gel electrophoresis patterns. J Appl Microbiol 97:446–458
Scheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40-base pair G + C rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci 86:232–235
Spano G, Lonvaud-Funel A, Claisse O, Massa S (2007) In vivo PCR-DGGE analysis of Lactobacillus plantarum and Oenococcus oeni populations in red wine. Curr Microbiol 54:9–13
Zapparoli G, Reguant C, Bordons A, Torriani S, Dellaglio F (2000) Genomic DNA fingerprinting of Oenococcus oeni strains by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA-PCR. Curr Microbiol 40:351–355
Acknowledgments
The authors wish to thank the Ministry of Education and Science (INIA-MEC) for its financial support (project RM 2006-00011-C02-02). P. Ruiz is supported by a grant from the Council of Communities of Castilla-La Mancha (JCCM).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ruiz, P., Seseña, S., Izquierdo, P.M. et al. Bacterial biodiversity and dynamics during malolactic fermentation of Tempranillo wines as determined by a culture-independent method (PCR-DGGE). Appl Microbiol Biotechnol 86, 1555–1562 (2010). https://doi.org/10.1007/s00253-010-2492-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2492-8