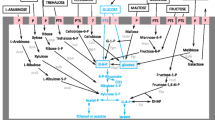
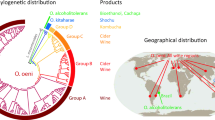
Abstract
Malolactic fermentation (MLF) is the bacterially driven decarboxylation of l-malic acid to l-lactic acid and carbon dioxide, and brings about deacidification, flavour modification and microbial stability of wine. The main objective of MLF is to decrease wine sourness by a small increase in wine pH via the metabolism of l-malic acid. Oenococcus oeni is the main lactic acid bacterium to conduct MLF in virtually all red wine and an increasing number of white and sparkling wine bases. Over the last decade, it is becoming increasingly recognized that O. oeni exhibits a diverse array of secondary metabolic activities during MLF which can modify the sensory properties of wine. These secondary activities include the metabolism of organic acids, carbohydrates, polysaccharides and amino acids, and numerous enzymes such as glycosidases, esterases and proteases, which generate volatile compounds well above their odour detection threshold. Phenotypic variation between O. oeni strains is central for producing different wine styles. Recent studies using array-based comparative genome hybridization and genome sequencing of three O. oeni strains have revealed the large genomic diversity within this species. This review will explore the links between O. oeni metabolism, genomic diversity and wine sensory attributes.


Similar content being viewed by others
References
Amerine MA, Roessler EB (1983) Wines—their sensory evaluation, 2nd edn. Freeman, San Francisco
Assad-Garcia JS, Bonnin-Jusserand M, Garmyn D, Guzzo J, Alexandre H, Grandvalet C (2008) An improved protocol for electroporation of Oenococcus oeni ATCC BAA-1163 using ethanol as immediate membrane fluidizing agent. Lett Appl Microbiol 47(4):333–338
Bartowsky EJ (2005) Oenococcus oeni and malolactic fermentation—moving into the molecular arena. Aust J Grape Wine Res 11(2):174–187
Bartowsky EJ, Henschke PA (1995) Malolactic fermentation and wine flavour. Aust Grapegrower Winemaker 378a:83–94
Bartowsky EJ, Henschke PA (2004) The ‘buttery’ attribute of wine—diacetyl—desirability, spoilage and beyond. Int J Food Microbiol 96:235–252
Bartowsky EJ, Henschke PA (2008) Acetic acid bacteria spoilage of bottled red wine—a review. Int J Food Microbiol 125:60–70
Bartowsky EJ, Pretorius IS (2008) Microbial formation and modification of flavour and off-flavour compounds in wine. In: König H, Unden G, Fröhlich J (eds) Biology of microorganisms on grapes, in must and wine. Springer, Heidelberg, pp 211–233
Bartowsky E, Costello P, Henschke P (2002a) Management of malolactic fermentation—wine flavour manipulation. Aust Grapegrower Winemaker 461a:7–12
Bartowsky EJ, Francis IL, Bellon JR, Henschke PA (2002b) Is buttery aroma perception in wines predictable from diacetyl concentration? Aust J Grape Wine Res 8:180–185
Bartowsky E, Costello P, McCarthy J (2008) MLF—adding an ‘extra dimension’ to wine flavour and quality. Aust NZ Grapegrower Winemaker 533a:60–65
Bartowsky E, Costello P, Krieger-Weber S, Markides A, Francis L, Travis B (2010) Influence of malolactic fermentation on the fruity characters of red wine—bringing wine chemistry and sensory together. In: International Intervitis Interfructa Congress 2010, Stuttgart, Germany, 24–28 March 2010. pp 52–59
Beltramo C, Oraby M, Bourel G, Garmyn D, Guzzo J (2004) A new vector, pGID052, for genetic transfer in Oenococcus oeni. FEMS Microbiol Lett 236(1):53–60
Bon E, Delaherche A, Bilhere E, De Daruvar A, Lonvaud-Funel A, Le Marrec C (2009) Oenococcus oeni genome plasticity is associated with fitness. App Environ Microbiol 75(7):2079–2090
Borneman AR, Bartowsky EJ, McCarthy J, Chambers PJ (2010) Genotypic diversity in Oenococcus oeni by high-density microarray comparative genome hybridization and whole genome sequencing. Appl Microbiol Biotechnol 86(2):681–691. doi:10.1007/s00253-009-2425-6
Canas PMI, Perez PR, Prieto SS, Herreros MLP (2009) Ecological study of lactic acid microbiota isolated from Tempranillo wines of Castilla-La Mancha. J Biosci Bioeng 108(3):220–224
Capaldo A, Walker ME, Ford CM, Jiranek V (2011) beta-Glucoside metabolism in Oenococcus oeni: cloning and characterisation of the phospho-beta-glucosidase bgID. Food Chem 125(2):476–482
Capozzi V, Russo P, Beneduce L, Weidmann S, Grieco F, Guzzo J, Spano G (2010) Technological properties of Oenococcus oeni strains isolated from typical southern Italian wines. Lett Appl Microbiol 50(3):327–334
Caspritz G, Radler F (1983) Malolactic enzyme of Lactobacillus plantarum. Purification, properties, and distribution among bacteria. J Biol Chem 258:4907–4910
Chambers PJ, Pretorius IS (2010) Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep 11(12):914–920
Cogan TM (1995) Flavour production by dairy starter cultures. J Appl Bacteriol Symp Suppl 79:49S–64S
Curtin CD, Bellon JR, Henschke PA, Godden P, de Barros LM (2007) Genetic diversity of Dekkera bruxellensis yeasts isolated from Australian wineries. FEMS Yeast Res 7(3):471–481
Dicks LMT (1994) Transformation of Leuconostoc oenos by electroporation. Biotechnol Tech 8(12):901–904
Dicks LMT, Dellaglio F, Collins MD (1995) Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int J Sys Evol Bacteriol 45(2):395–397
D'Incecco N, Bartowsky E, Kassara S, Lante A, Spettolli P, Henschke P (2004) Release of glycosidically bound flavour compounds of Chardonnay by Oenococcus oeni during malolactic fermentation. Food Microbiol 21(3):257–265
Endo A, Okada S (2006) Oenococcus kitaharae sp nov., a non-acidophilic and non-malolactic-fermenting oenococcus isolated from a composting distilled shochu residue. Int J Sys Evol Microbiol 56:2345–2348. doi:10.1099/ijs.0.64288-0
Eom HJ, Cho SK, Park MS, Ji GE, Han NS (2010) Characterization of Leuconostoc citreum plasmid pCB18 and development of broad host range shuttle vector for lactic acid bacteria. Biotechnol Bioprocess Eng 15(6):946–952. doi:10.1007/s12257-010-0089-9
Francis IL, Newton JL (2005) Determining wine aroma from compositional data. Aust J Grape Wine Res 11(2):114–126
Garmyn D, Monnet C, Martineau B, Guzzo J, Cavin J-F, Diviès C (1996) Cloning and sequencing of the gene encoding α-acetolactate decarboxylase from Leuconostoc oenos. FEMS Microbiol Lett 145:445–450
Garvie EI (1967) Leuconostoc oenos sp.nov. J Gen Microbiol 48(3):431–438. doi:10.1099/00221287-48-3-431
Gerbaux V, Briffox C, Dumont A, Krieger S (2009) Influence of inoculation with malolactic bacteria on volatile phenols in wines. Am J Enol Vitic 60(2):233–235
Grimaldi A, McLean H, Jiranek V (2000) Identification and partial characterization of glycosidic activities of commercial strains of the lactic acid bacterium, Oenococcus oeni. Am J Enol Vitic 51(4):362–369
Grimaldi A, Bartowsky E, Jiranek V (2005) A survey of glycosidase activities of commercial wine strains of Oenococcus oeni. Int J Food Microbiol 105(2):233–244
Gunata Z, Bitteur S, Brillouet J-M, Bayonove C, Cordonnier R (1988) Sequential enzymatic hydrolysis of potentially aromatic glycosides from grape. Carbohyd Res 184:139–149
Gunata YZ, Bayonove CL, Tapiero C, Cordonnier RE (1990) Hydrolysis of grape monoterpenyl β-D-glucosides by various β-glucosidases. J Agric Food Chem 38:1232–1236
Guth H (1997a) Identification of character impact odorants of different white wine varieties. J Agric Food Chem 45(8):3022–3026
Guth H (1997b) Quantitation and sensory studies of character impact odorants of different white wine varieties. J Agric Food Chem 45(8):3027–3032
Henick-Kling T (1993) Malolactic fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic Publisher, Amsterdam, pp 289–326
Iland P, Gago P, Caillard A, Dry P (2009) A taste of the world of wine. Patrick Iland Wine Promotions Pty Ltd, Campbelltown
Kunkee RE (1967) Malo-lactic fermentation. Adv Appl Microbiol 9:235–279
Kunkee RE (1991) Some roles of malic acid in the malolactic fermentation in wine making. FEMS Microbiol Rev 88:55–72
Labarre C, Divies C, Guzzo J (1996a) Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. App Environ Microbiol 62(12):4493–4498
Labarre C, Guzzo J, Cavin JF, Divies C (1996b) Cloning and characterization of the genes encoding the malolactic enzyme and the malate permease of Leuconostoc oenos. App Environ Microbiol 62(4):1274–1282
Laurent M-H, Henick-Kling T, Acree TE (1994) Changes in the aroma and odor of Chardonnay wine due to malolactic fermentation. Vitic Enol Sci 49:3–10
Li H, Zhang CH, Liu YL (2006) Species attribution and distinguishing strains of Oenococcus oeni isolated from Chinese wines. World J Microb Biot 22(5):515–518
Lonvaud-Funel A (2001) Biogenic amines in wines: role of lactic acid bacteria. FEMS Microbiol Lett 199(1):9–13
Lonvaud-Funel A, Strasser de Saad AM (1982) Purification and properties of a malolactic enzyme from a strain of Leuconostoc mesenteroides isolated from grapes. App Environ Microbiol 43:357–361
Loureiro V, Malfeito-Ferreira M (2003) Spoilage yeasts in the wine industry. Int J Food Microbiol 86:23–50
Martineau B, Henick-Kling T (1995) Performance and diacetyl production of commercial strains of malolactic bacteria in wine. J Appl Bacteriol 78:526–536
Martineau B, Acree TE, Henick-Kling T (1995) Effect of wine type on the detection threshold for diacetyl. Food Res Int 28(2):139–143
Matthews A, Grimaldi A, Walker M, Bartowsky E, Grbin P, Jiranek V (2004) Lactic acid bacteria as a potential source of enzymes for use in vinification. App Environ Microbiol 70(10):5715–5731. doi:10.1128/aem.70.10.5715-5731.2004
Michlmayr H, Schumann C, da Silva N, Kulbe KD, del Hierro AM (2010a) Isolation and basic characterization of a beta-glucosidase from a strain of Lactobacillus brevis isolated from a malolactic starter culture. J Appl Microbiol 108(2):550–559. doi:10.1111/j.1365-2672.2009.04461.x
Michlmayr H, Schumann C, Wurbs P, da Silva N, Rogl V, Kulbe KD, del Hierro AM (2010b) A beta-glucosidase from Oenococcus oeni ATCC BAA-1163 with potential for aroma release in wine: cloning and expression in E. col. World J Microb Biot 26(7):1281–1289
Michlmayr H, Schumann C, Kulbe KD, del Hierro AM (2011) Heterologously expressed family 51 α-L-arabinofurnaosidases from Oenococcus oeni and Lactobacillus brevis. App Environ Microbiol 77(4):1528–1531
Mills DA, Rawsthorne H, Parker C, Tamir D, Makarova K (2005) Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking. FEMS Microbiol Rev 29:465–475
Möslinger (1901) Über die Säuren des Weines und den Säurerückgang. Zeitschr f Untersuchung d Nahr Genussmittel 4:1120–1130
Müller-Thurgau H (1891) Über die Ergebnisse neuer Untersuchungen auf den Gebiete der Weinbereitung. Ber XII Dtsch Weinbaukong (Worms):128
Müller-Thurgau H, Osterwalder A (1913) Die Bakterien im Wein und Obstwein und die dadurch verursachten Veränderungen. Zentr Bakt Parasitenk Hyg Abt 2(36):129–338
Naouri P, Chagnaud P, Arnaud A, Galzy P (1990) Purification and properties of a malolactic enzyme from Leuconostoc oenos ATCC-23278. J Basic Microb 30(8):577–585
Nielsen JC, Prahl C, Lonvaud-Funel A (1996) Malolactic fermentation in wine by direct inoculation with freeze-dried Leuconostoc oenos cultures. Am J Enol Vitic 47(1):42–48
Pasteur L (1873) Études sur le Vin, 2nd edn. Savy, Paris
Pineau B, Barbe JC, Van Leeuwen C, Dubourdieu D (2009) Examples of perceptive interactions involved in specific “red-” and “black-berry” aromas in red wines. J Agric Food Chem 57(9):3702–3708
Pineau B, Barbe JC, van Leeuwen C, Dubourdieu D (2010) Olfactory specificity of red- and black-berry fruit aromas in red wines and contribution to the red Bordeaux wine concept. Journal International Des Sciences De La Vigne Et Du Vin 44(1):39–49
Ramos A, Poolman B, Santos H, Lolkema JS, Konings WN (1994) Uniport of anionic citrate and proton consumption in citrate metabolism generates a proton motive force in Leuconostoc oenos. J Bacteriol 176(16):4899–4905
Ramos A, Lolkema JS, Konings WN, Santos H (1995) Enzyme basis for pH regulation of citrate and pyruvate metabolism by Leuconostoc oenos. App Environ Microbiol 61(4):1303–1310
Renouf V, Claisse O, Lonvaud-Funel A (2007) Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol 75(1):149–164
Ruiz P, Izquierdo PM, Sesena S, Palop ML (2010) Selection of autochthonous Oenococcus oeni strains according to their oenological properties and vinification results. Int J Food Microbiol 137(2–3):230–235
Schmid F, Li Y, Liebich B, Culbert J, Day C, Jiranek V (2007) Evaluation of red wine made on a small scale utilizing frozen must. J Agric Food Chem 55(17):7156–7161
Segurel MA, Razungles AJ, Riou C, Salles M, Baumes RL (2004) Contribution of dimethyl sulfide to the aroma of Syrah and Grenache Noir wines and estimation of its potential in grapes of these varieties. J Agric Food Chem 52(23):7084–7093
Sico MA, Bonomo MG, Salzano G (2008) Isolation and characterization of Oenococcus oeni from Aglianico wines. World J Microb Biot 24(9):1829–1835
Siebert TE, Smythe HE, Capone DL, Neuwöhner C, Pardon KH, Skouroumounis GK, Herderich MJ, Sefton MA, Pollnitz AP (2005) Stable isotope dilution analysis of wine fermentation products by HS = SPME-GC-MS. Anal Bioanal Chem 381:937–947
Smit G, Smit BA, Engels WJM (2005) Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev 29(3):591–610
Solieri L, Genova F, De Paola M, Giudici P (2010) Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures. J Appl Microbiol 108(1):285–298
Spettoli P, Nuti MP, Zamorani A (1984) Properties of malolactic activity purified from Leuconostoc oenos ML34 by affinity chromatography. App Environ Microbiol 9:184–189
Sponholz W-R (1993) Wine spoilage by microorganisms. In: Fleet GH (ed) Wine microbiology and technology. Harwood Academic Publishing, Amsterdam, pp 395–420
Swiegers JH, Bartowsky EJ, Henschke PA, Pretorius IS (2005) Yeast and bacterial modulation of wine aroma and flavour. Aust J Grape Wine Res 11(2):139–173
Ugliano M, Moio L (2005) Changes in the concentration of yeast-derived volatile compounds of red wine during malolactic fermentation with four commercial starter cultures of Oenococcus oeni. J Agric Food Chem 53(26):10134–10139
Ugliano M, Moio L (2006) The influence of malolactic fermentation and Oenococcus oeni strain on glycosidic aroma precursors and related volatile compounds of red wine. J Sci Food Agric 86(14):2468–2476. doi:10.1002/jsfa.2650
Ugliano M, Genovese A, Moio L (2003) Hydrolysis of wine aroma precursors during malolactic fermentation with four commercial starter cultures of Oenococcus oeni. J Agric Food Chem 51(17):5073–5078. doi:10.1021/jf0342019
Vigentini I, Picozzi C, Tirelli A, Giugni A, Foschino R (2009) Survey on indigenous Oenococcus oeni strains isolated from red wines of Valtellina, a cold climate wine-growing Italian area. Int J Food Microbiol 136(1):123–128
Williams PJ, Strauss CR, Wilson B, Massy-Westropp RA (1982) Novel monoterpene disaccharide glycosides of Vitis vinifera grapes and wines. Phytochemistry 21:2013–2020
Williams PJ, Sefton MA, Wilson B (1989) Nonvolatile conjugates of secondary metabolites as precursors of varietal grape flavour components. In: Teranishi R, Buttery RG, Shahisi R (eds) Flavour chemistry, trends and developments, ACS symposium series no 388. American Chemical Society, Washington DC, pp 35–48
Yanagida F, Srionnual S, Chen YS (2008) Isolation and characteristics of lactic acid bacteria from koshu vineyards in Japan. Lett Appl Microbiol 47(2):134–139. doi:10.1111/j.1472-765X.2008.02398.x
Ze-Ze L, Teneiro R, Brito L, Santos MA, Paveia H (1998) Physical map of the genome of Oenococcus oeni PSU-1 and localization of genetic markers. Microbiology 144:1145–1156
Ze-Ze L, Teneiro R, Paveia H (2000) The Oenococcus oeni genome: physical and genetic map of strain GM and comparison with the genome of a ‘divergent’ strain, PSU-1. Microbiology 146(12):3195–3204
Ze-Ze L, Chelo IM, Tenreiro R (2008) Genome organization in Oenococcus oeni strains studied by comparison of physical and genetic maps. Int Microbiol 11(4):237–244
Acknowledgements
The authors thank Dr. Paul Chambers for discussions during the preparation of this mini review and Lallemand SA for allowing us to use data presented in Fig. 2 from a project for which they provided financial support. The Australian Wine Research Institute (AWRI) is supported by Australian grapegrowers and winemakers through their investment agency, the Grape and Wine Research and Development Corporation, with matching funds from the Australian Government. The AWRI is a member of the Wine Innovation Cluster in Adelaide.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bartowsky, E.J., Borneman, A.R. Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine. Appl Microbiol Biotechnol 92, 441–447 (2011). https://doi.org/10.1007/s00253-011-3546-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3546-2