Abstract
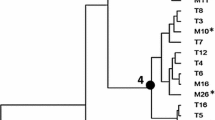
The aim of this study was to evaluate fifty-three Lactobacillus plantarum isolates obtained from a Patagonian red wine, molecularly identified and typified using RAPD analysis, in order to select starter cultures for malolactic fermentation (MLF). The results obtained suggest a considerable genetic diversity, taking into account that all L. plantarum isolates were obtained from one cellar and one vintage. Based on the capacity to tolerate a concentration of 14 % ethanol in MRS broth for 2 days, eight isolates were selected for the subsequent analysis. The incidence of various wine stress factors (ethanol, acid pH, lysozyme and sulfur dioxide) on isolates growth was studied. Besides, glucosidase and tannase activities were evaluated, and the presence of genes involved in the synthesis of biogenic amines was examined by PCR. A previously characterized indigenous Oenococcus oeni strain was included with comparative purposes. Differences in technologically relevant characteristics were observed among the eight L. plantarum selected isolates, revealing an isolate-dependent behavior. Detectable glucosidase and tannase activities were found in all isolates. The presence of genes encoding histidine and tyrosine descarboxylases and putrescine carbamoyltransferase was not detected. The ability of L. plantarum isolates to grow and consume l-malic acid in simulated laboratory-scale vinifications revealed that two of them could be considered as possible MLF starter cultures for Patagonian red wines. These isolates will be subjected to further analysis, for a final winery technological characterization.






Similar content being viewed by others
References
Apostol BL, Black WC IV, Miller BR, Reiter P, Beaty BJ (1993) Estimation of the number of full sibling families at an oviposition site using RAPD—PCR markers: applications to the mosquito Aedes aegypti. Theor Appl Genet 86(8):991–1000
Bae S, Fleet GH, Heards GM (2006) Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100(4):712–727
Bartowsky EJ, Henschke PA (2004) The buttery attribute of wine—diacetyl—desirability, spoilage and beyond. Int J Food Microbiol 96(3):235–252
Bauer R, Dicks LMT (2004) Control of malolactic fermentation in wine. A review. S Afr J Enol Vitic 25(2):74–88
Bravo-Ferrada BM, Delfederico L, Hollmann A, La Valdés HensD, Curilén Y, Caballero A, Semorile L (2011) Oenococcus oeni from patagonian red wines: isolation, characterization and technological properties. Int J Microbiol Res 3(1):48–55
Cappozzi V, Russo P, Beneduce L, Weidmann S, Grieco F, Guzzo J, Spano G (2010) Technological properties of Oenococcus oeni strains isolated from typical southern Italian wines. Lett Appl Microbiol 50(3):327–334
Carreté R, Reguant C, Rozés N, Constantí M, Bordons A (2006) Analysis of Oenococcus oeni strains in simulated microvinifications with some stress compounds. Am J Enol Vitic 57(3):356–362
Cho GS, Krauss S, Huch M, du Toit M, Franz CM (2011) Development of a quantitative PCR for detection of Lactobacillus plantarum starters during wine malolactic fermentation. J Microbiol Biotechnol 21(12):1260–1266
Chu-Ky S, Tourdot-Marechal R, Marechal PA, Guzzo J (2005) Combined cold, acid, ethanol shocks in Oenococcus oeni: effects on membrane fluidity and cell viability. Biochim Biophys Acta 1717(2):118–124
Claisse O, Renouf V, Lonvaud-Funel A (2007) Differentiation of wine LAB species based on RFLP analysis of a partial sequence of rpoB gene. J Microbiol Methods 69(2):387–390
Cocconcelli PS, Porro D, Galandini S, Senini L (1995) Development of RAPD protocol for typing of strains of LAB and enterococci. Lett Appl Microbiol 21(6):376–379
Davis CR, Wibowo D, Eschenbruch R, Lee TH, Fleet GH (1985) Practical implications of malolactic fermentation: a review. Am J Enol Vitic 36(4):290–301
Delfederico L, Hollmann A, Martínez M, Iglesias NG, De Antoni G, Semorile L (2006) Molecular identification and typing of lactobacilli isolated from kefir grains. J Dairy Res 73(1):20–27
Delfini C, Morsiani MG (1992) Resistance to sulfur dioxide of malolactic strains of Leuconostoc oenos and Lactobacillus sp isolated from wines. Sciences des Aliments 12(3):493–511
Delfini C, Cersosimo M, del Prete V, Strano M, Gaetano G, Pagliara A, Ambró S (2004) Resistance screening assay of wine lactic acid bacteria on lysoszyme: efficacy of lysozyme in unclarified grape musts. J Agric Food Chem 52(7):1861–1866
Du Plessis HW, Dicks LMT, Pretorius IS, Lambrechts MG, du Toit M (2004) Identification of lactic acid bacteria isolated from South African brandy base wines. J Food Microbiol 91(1):19–29
du Toit M, Englebrecht L, Lerm E, Krieger-Weber S (2011) Lactobacillus: the next generation of malolactic starter cultures—an overview. Food Bioproc Technol 4(6):876–906
G-Alegría E, López I, Ignacio RuizJ, Sáenz J, Fernández E, Zarazaga M, Dizy M, Torres C, Ruiz-Larrea F (2004) High tolerance of wild Lactobacillus plantarum and Oenococcus oeni strains to lyophilisation and stress environmental conditions of acid pH and ethanol. FEMS Microbiol Lett 230(1):53–61
García Barceló J (1990) Dióxido de azufre. In: Técnicas analíticas para vinos, 1st edn. GAB, Barcelona, España, pp 3.5–3.10
Gindreau E, Keim H, de Revel G, Bertrand A, Lonvaud-Funel A (2003) Use of direct inoculation malolactic starters: setting efficiency and sensorial impact. J Int Sci Vig Vin 37:51–57
Gockowick H, Henschke PA (2003) Interaction of pH, ethanol concentration and wine matrix on induction of malolactic fermentation with commercial direct inoculation starter culture. Austral J Grape Wine Res 9(3):200–209
Gónzalez-Arenzana L, López R, Santamaría P, Tenorio C, López-Alfaro I (2012) Dynamics of indigenous lactic acid bacteria populations in wine fermentations from La Rioja (Spain) during three vintages. Environ Microbiol 63(1):12–19
Grimaldi A, McLean H, Jiranek V (2000) Identification and partial characterization of glycosidic activities of commercial strains of the lactic acid bacteria Oenococcus oeni. Am J Enol Vitic 51(4):362–369
Henick-Kling T, Sandine WE, Heatherbell DA (1989) Evaluation of malolactic bacteria isolated from Oregon wines. Appl Environ Microbiol 55(8):2010–2016
Kempler GM, McKay LL (1980) Improved medium for detection of citrate-fermenting Streptococcus lactis subsp diacetylactis. Appl Environm Microbiol 39(4):926–927
Knoll C, Divol B, du Toit M (2008) Genetic screening of lactic acid bacteria of oenological origin for bacteriocin-encoding genes. Food Microbiol 25(8):983–991
Landete JM, Arena ME, Pardo I, Manca de Nadra MC, Ferrer S (2010) The role of two families of bacterial enzymes in putrescine synthesis from agmatine via agmatine deiminase. Int Microbiol 13(4):169–177
Le Jeune C, Lonvaud-Funel A, Ten Brink B, Hofstra H, van der Vossen JMBM (1995) Development of a detection system for histidine decarboxilating lactic acid bacteria based on DNA probes, PCR and activity test. J Appl Bacteriol 78(3):316–326
Lee SG, Lee KW, Park TH, Park JY, Han NS, Kim JH (2012) Proteomic analysis of proteins increased or reduced by ethanol of Lactobacillus plantarum ST4 isolated from Makgeoli, traditional Korean rice wine. J Microbiol Biotechnol 22(4):516–525
Lekha PK, Lonsane BK (1997) Production and application of tannin acyl hydrolase: state of the art. Adv Appl Microbiol 44:215–260
Lerm E, Engelbrecht L, du Toit M (2011) Selection and characterization of Oenococcus oeni and Lactobacillus plantarum South African wine isolates for use as malolactic fermentation starter cultures. S Afr J Enol Vitic 32(2):280–295
Liu SQ (2002) A review: Malolactic fermentation in wine—beyond deacidification. J Appl Microbiol 92(4):589–601
Lonvaud-Funel A (1999) Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 76(1):317–331
Lopes CA, Rodríguez ME, Sangorrín M, Querol A, Caballero AC (2007) Patagonian wines: the selection of an indigenous yeast starter. J Indus Microbiol Biotechnol 34(8):539–546
López I, López R, Santamaría P, Torres C, Ruiz-Larrea F (2008) Performance of malolactic fermentation by inoculation of selected Lactobacillus plantarum and Oenococcus oeni strains isolated from Rioja red wines. Vitis 47(2):123–129
Lucas P, Lonvaud-Funel A (2002) Purification and parcial gene sequence of the tyrosine decarboxylase of Lactobacillus brevis IOEB 9809. FEMS Microbiol Lett 211(1):85–89
Lucas P, Landete J, Coton M, Coton E, Lonvaud-Funel A (2003) The tyrosine decarboxylase operon of Lactobacillus brevis IOEB 9809: characterization and conservation in tyramine-producing bacteria. FEMS Microbiol Lett 229(1):65–71
Maicas S, Gil JV, Pardo I, Ferrer S (1999) Improvement of volatile composition of wines by controlled addition of malolactic bacteria. Food Res Int 32(7):491–496
Miller BJ, Franz CM, Cho GS, du Toit M (2011) Expression of the malolactic enzyme gene (mle) from Lactobacillus plantarum under winemaking conditions. Curr Microbiol 62(6):1682–1688
Moreno-Arribas MV, Torlois S, Joyeux A, Bertrand A, Lonvaud-Funel A (2000) Isolation, properties ad behaviour of tyramine-producing lactic acid bacteria from wine. J Appl Microbiol 88(4):584–593
Moreno-Arribas MV, Polo CM, Jorganes F, Muñoz R (2003) Screening of biogenic amine production by lactic acid bacteria isolated from grape must and wine. Int J Food Microbiol 84(1):117–123
Osawa R, Walsh TP (1993) Visual reading method for detection of bacterial tannase. Appl Environ Microbiol 59(4):1251–1252
Parente E, Ciocia F, Ricciardi A, Zotta T, Felis GE, Torriani S (2010) Diversity of stress tolerance in Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus paraplantarum: a multivariate screening study. Int J Food Microbiol 144(2):270–279
Reglamento CEE Nº 2676/90 (1990) In: Diario Oficial de las Comunidades Europeas, pp 123–130
Rodas AM, Ferrer S, Pardo I (2003) 16S-ARDRA, a tool for identification of lactic acid bacteria isolated from grape must and wine. Syst Appl Microbiol 26(3):412–422
Rojo-Bezares B, Sáenz Y, Navarro L, Zarazaga M, Ruiz-Larrea F, Torres C (2007) Co-culture inducible bacteriocin activity of Lactobacillus plantarum strain J23 isolated from grape must. Food Microbiol 24(5):482–491
Ruiz P, Izquierdo PM, Seseña S, Llanos Palop M (2008) Intraspecific genetic diversity of lactic acid bacteria from malolactic fermentation of Cencibel wines as derived from combined analysis of RAPD-PCR and PFGE patterns. Food Microbiol 25(7):942–948
Ruiz P, Izquierdo PM, Seseña S, Llanos Palop M (2010) Selection of autochthonous Oenococcus oeni strains according to their oenological properties and vinification results. Int J Food Microbiol 137(2):230–235
Sestelo ABF, Poza M, Villa TG (2004) β-glucosidase of Lactobacillus plantarum wine strain. Worl J Microbiol Biotechnol 20(6):633–637
Sico MA, Bonomo MG, Salzano G (2008) Isolation and characterization of Oenococcus oeni from Aglianico wines. Worl J Microbiol Biotechnol 24(9):1829–1835
Solieri L, Genova F, De Paola M, Giudici P (2009) Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures. J Appl Microbiol 108(1):285–298
Ulrike E, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene encoding for 16S ribosomal RNA. Nucl Acid Res 17(19):7843–7853
Vaquero I, Marcobal A, Muñoz R (2004) Tannase activity by lactic acid bacteria isolated from grape must and wine. Int J Food Microbiol 96(2):199–204
Acknowledgments
This work was supported by grants from Universidad Nacional de Quilmes (Programa Microbiología Molecular Básica y Aplicaciones Biotecnológicas and ANPCyT—PICTO UNQ 2006 Nº 36474). Bravo Ferrada BM was recipient of a fellowship from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). Dr. L. Semorile is member of the Research Career of Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CIC-PBA)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bravo-Ferrada, B.M., Hollmann, A., Delfederico, L. et al. Patagonian red wines: selection of Lactobacillus plantarum isolates as potential starter cultures for malolactic fermentation. World J Microbiol Biotechnol 29, 1537–1549 (2013). https://doi.org/10.1007/s11274-013-1337-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-013-1337-x