Abstract
Alzheimer’s disease (AD) pathogenesis feature progressive neurodegeneration, amyloid-β plaque formation, and neurofibrillary tangles. Ample evidence has indicated the involvement of epigenetic pathways in AD pathogenesis. Here, we show that the expression of microRNA 650 (miR-650) is altered in brains from AD patients. Furthermore, we found that the processing of primary miR-650 to mature miR-650 is misregulated. Bioinformatic analysis predicted that miR-650 targets the expression of three AD-associated components: Apolipoprotein E (APOE), Presenilin 1 (PSEN1), and Cyclin-Dependent Kinase 5 (CDK5), and we have experimentally confirmed that miR-650 is able to significantly reduce the expression of APOE, PSEN1, and CDK5 in vitro. Importantly, the overexpression of miR-650 was further shown to significantly alter the CDK5 level and ameliorate AD pathologies in APP-PSEN1 transgenic mice. Overall, our results indicate that miR-650 influences AD pathogenesis through regulation of CDK5.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is the most common form of neurodegenerative diseases, and the leading cause of dementia. Although the etiology of AD is still rather vague, multiple neuropathological hallmarks of AD have been characterized, including insoluble, extracellular aggregates of amyloid-β peptides (Aβ), named amyloid plaques, and intracellular accumulations of hyperphosphorylated microtubule-associated protein tau, named neurofibrillary tangles (NFTs), as well as neuronal loss [1,2,3,4]. At the molecular level, tremendous efforts have been made to dissect various pathways and components underlying the pathogenesis of neurodegenerative lesions observed in AD. The processing of amyloid precursor protein (APP) to Aβ is catalyzed by β-secretase and γ-secretase complex sequentially, and the resultant cleaved short forms of Aβ can then self-aggregate to form amyloid plaques. It is widely accepted that the amyloidogenesis results in the activation of various kinases, leading to the neurotoxic phosphorylation of tau [5]. Nonetheless, the molecular basis, especially at the early onset of AD, is still unclear.
The interactions between protein kinase and phosphatase activities, via switching on and off of protein phosphorylation, regulate numerous cellular processes. Indeed, abnormally phosphorylated proteins have been conclusively documented in various diseases, including degenerative diseases in the brain [6,7,8,9]. In particular, several kinases that regulate the production of Aβ peptide, as well as tau phosphorylation, were altered in AD patients [10,11,12,13,14,15,16]. For instance, inhibition of CDK5 appears to protect β-amyloid-induced neuronal cell death; treatment with therapeutic concentration of a glycogen synthase kinase inhibitor blocked the generation of β-amyloid peptides. However, little is known about the regulating mechanisms of the related kinase expression in the context of AD pathogenesis.
As one component of the epigenetic landscape, microRNAs (miRNAs) are small non-coding RNAs with a length of ~ 22 nucleotides (nt). The biogenesis of miRNA is a multi-step process [17,18,19,20]. Transcribed primary miRNAs (pri-miRNAs) are processed by Drosha in the nucleus, and the resulting precursor miRNAs (pre-miRNAs) are exported to the cytoplasm where they are further processed by Dicer to generate mature miRNAs. The produced miRNAs regulate gene expression through binding of the 3′-untranslated regions (3′-UTR) of target mRNAs. Previous studies have shown that miRNAs play essential functions in a diversity of biological processes including cancer, development, aging, and disease progression [21,22,23,24,25,26]. In neuronal systems, previous studies have demonstrated that miRNAs regulate neuronal development and synaptic plasticity, toxic protein accumulation, and cell death and survival, suggesting their involvement in neuronal degenerative diseases [22, 25, 27]. The aberrant miRNA levels have been indicated in the pathogenesis of neurodegenerative diseases including Parkinson’s disease and Alzheimer’s disease [28,29,30,31,32,33,34,35]. Nonetheless, how the deregulated miRNAs might contribute to disease pathogenesis is still not fully understood.
To gain more insight into the function of miRNAs in the pathogenesis of AD, we profiled the miRNA expression in the brain of normal and AD patients and uncovered several miRNAs that showed a significant alteration among AD cases. Using bioinformatic analyses, we determined that miR-650, one of these altered miRNAs, was likely to bind to APOE, PSEN1, and CDK5, genes involved in AD pathogenesis. We further found that the process from primary/precursor to the mature form of miR-650 was dysregulated in AD cases. Importantly, we discovered that miR-650 can regulate APOE, PSEN1, and CDK5 expression levels in vitro, and we went on to determine that miR-650 could also regulate CDK5 levels in vivo using APP/PSEN1 mice, a conventional model for AD. Over-expression of miR-650 could also ameliorate neuronal deficits in these AD mice as evidenced by a decrease in the number of plaques and decreased Aβ levels. Our studies indicate that miR-650-CDK5 axis plays a regulatory role in the pathogenesis of AD.
Materials and Methods
Patient Information
Brain tissues of control and AD patients were obtained from archival human brain specimens of the Emory ADRC Neuropathology Core, using appropriate IRB protocols. Neuropathologic examination of every subject was recorded, and patients were diagnosed according to the National Institute on Aging/Alzheimer’s Association (NIA/AA) criteria [36]. Criteria for inclusion are the following: (1) post-mortem interval < = 8 h; (2) patient aged 65 and older; (3) no complicating agonal condition or underlying CNS disease, including stroke, neoplasms, or features of non-AD dementia (cortical Lewy bodies); and (4) extensive premortem cognitive testing (followed at least 3 years, with a battery of tests administered within one year of death). In order to provide adequate inter- and intra-case controls, tissues must be sampled from different cerebral cortical areas. The inclusion of an age-matched cohort with no cognitive impairment as a control group for comparison with the AD cases is also essential. Superior frontal (Brodmann area 9) and paracalcarine cortex (Brodmann area 17) from both controls and patients were collected by the Emory ADRC Neuropathology Core and used for RNA isolation. The information of patients and healthy controls was summarized in Table 1.
APP/PSEN1 Double Transgenic Mice
Age-matched male wild-type (WT) and APP/PSEN1 double transgenic littermate mice were used in this study. APP/PSEN1 double transgenic mice expressing a chimeric mouse/human APP with the Swedish mutation and a human PSEN1 Δexon 9 mutation [37, 38] were obtained from The Jackson Laboratory. Mice were maintained at ambient temperature (22–24 °C) on a 12:12 light/dark cycle with free access to food and water. All animal procedures were performed according to protocols approved by Emory University Institutional Animal Care and Use Committee.
Relative Quantification of Mature miRNAs by TaqMan Assay
Profiling of mature miRNA expression was performed using TaqMan miRNA assays (Applied Biosystems) as described previously [39]. In brief, 48 reverse transcription primers were used in 20-µl reactions consisting of: 20 ng total RNA, 1 × TaqMan miRNA reverse transcription primer pool, 0.5 mM of each deoxyribonucleotide triphosphate (dNTP), 10.0 U/µl MultiScribe (Applied Biosystems) reverse transcription, 1 × reverse transcription buffer, 0.25 U/µl RNase inhibitor, and nuclease-free water. The reactions were incubated at 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. Reactions were diluted 1:10 with nuclease-free water for use in the TaqMan real-time PCR reactions. Individual TaqMan miRNA real-time PCR reactions for profiling experiments were performed on a PCR system (7900HT SDS; Applied Biosystems) in a 384-well format running control and AD samples in parallel for each cDNA pool generated in the reverse transcription step. PCR reactions were performed in triplicate for each sample and each miRNA. The 10-µl reactions consisted of 1 × TaqMan Universal Master Mix, No AmpErase UNG, 1 × TaqMan miRNA assay mix, 0.8 µl of 1:10 diluted cDNA, and nuclease-free water. All TaqMan PCR reactions were prepared and aliquoted using a custom method on an automated pipette (Biomek FX; Beckman Coulter). PCR reaction conditions were run according to the standard protocol without the 50 °C incubation using version 2.3 of the SDS software, with reactions incubated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min.
For the reverse transcription of specific miRNA, the 15-μl reverse transcription reaction consisted of 1 μg of total RNA, 1 × of specific RT primer, 5 U MultiScribe Reverse transcription, and 0.5 mM of dNTPs mixture, 1 × reverse transcription buffer, 4 U RNase inhibitor, and nuclease-free water. Reverse transcription reactions were performed as followings: 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min for the TaqMan assay of individual miRNAs.
Primary/precursor miR-650 was detected by polyadenylating total RNA using SuperScript III oligo-dT reverse transcription (Invitrogen) to generate first-strand cDNA and real-time PCR targeting the pri-/pre-miR-650. Polyadenylation and reverse transcription were performed according to the manufacturer’s instructions using the Ncode miRNA First-Strand cDNA Synthesis kit (Invitrogen), with 1 µg of total RNA isolated with TRIZOL as input. Relative quantification was performed by real-time PCR using 1:10 diluted cDNA.
Plasmid Construction, Co-transfection, and Luciferase Assay
To construct the 3′-UTR-luciferase vectors, the 3′-UTR sequence of APOE, PSEN1, and CDK5 mRNA were PCR amplified from 293FT cell cDNA. The following primers were used: APOE: 3′-UTR–F: TCAGGAGCTCACGCCGAAGCCTGCAGCCATG, 3′-UTR–R: TCAGTCTAGATGCGTGAAACTTGGTGAATCT; PSEN1: 3′-UTR–F: TCAGGAGCT CC ATATTTGCGGTTAGAATCCCATG, 3′-UTR–R: TCAGTCTAGATGCTTCAACAGCCA TTTTACTCTT; and CDK5:3′-UTR–F: TCAGGAGCTCGCCCCGGGACCCCCGGCCT CCAGG, 3′-UTR–R: TCAGTCTAGAGACTGTGGGAAAG GAGCCAATTTA. The PCR products were each cloned into a pIS2 vector (Addgene) at SacI and XbaI sites.
To construct the mutant plasmids, the miR-650 target site in the 3′-UTRs of APOE, PSEN1, and CDK5 were deleted, using the QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technologies, Cat# 200514). The following primers were used for mutagenesis: mutant mutant APOE: CGCTGCAGGCTGCGCCGGGGTGGCGTGGGG; PSEN1: GGGAGAGA AGAATGAGGACAGTCTGGGGCAAGTGAGCGTACA; CDK5: TCTAGAGACTGT GGGAAATTTATGAAATTAAATAAAGTCCAC. The mutant plasmids were verified by sequencing.
For the transfection, pIS2-3′UTR (0.4 μg) and pIS0 (0.4 μg) were co-transfected with miR-650 duplex at a final concentration of 2.5 nM into 293FT cells with Lipofectamine 2000 (Invitrogen, Cat#11668019) in 24-well plate. pIS0 vector (Addgene) was used for the internal control. The dual luciferase assay was performed according to the manufacturer’s instructions (Promega, Cat# TM040) 48 h after the transfection. All assays were performed in triplicate and repeated independently three times for each co-transfection.
Human miR-650 precursor sequence was fused with a GFP tag in the N terminus and subcloned into pAAV2/9n vector (Addgene). The CMV promoter sequence was inserted into the construct to replace the original promoter in the vector. AVV-GFP-miR-650 and AAV-GFP (used as a control) was packaged and amplified by the Viral Vector Core at Emory University.
AAV Virus In Vivo Injection
The in vivo virus grafting was performed as described previously [39]. Briefly, 1-year-old APP/PSEN1 double transgenic mice were anesthetized with 1% isoflurane, and AAV virus (3ul with titer: 8.0X1013 VG/ml) was injected stereotaxically into the dentate gyrus (DG) using the following coordinates relative to the bregma: anteroposterior, − (1/2) × d mm; lateral, ± 1.8 mm (if d > 1.6), or otherwise ± 1.7 mm; and ventral, − 1.9 mm (from dura). For each mouse, the AAV-GFP control virus was injected into the left DG, and the AAV-miR-650 virus was injected into the right DG. The mice were sacrificed, and hippocampi were dissected out 1 month after the virus grafting for biochemical assay.
Western Blot Analyses
The tissues were ground in ProteoJET Mammalian Cell Lysis Reagent (Fermentas, Cat#K0301) with the presence of protease inhibitor cocktail (Roche, Cat# 11836153001). Lysate was kept on ice for 30 min. Thirty-microgram total protein were separated on SDS-PAGE gels and then transferred to PVDF membranes (Millipore). Membranes were processed following the HyGLO QuickSpray western blotting protocol (Denville). The following antibodies were used: polyclonal anti-Cdk5 (C-8) (Santa Cruz, Cat# sc-173), polyclonal anti-GFP (Invitrogen, Cat#A11122), and monoclonal anti-human GAPDH (Invitrogen, Cat# 437000). The density of each band was quantified by ImageJ software. An unpaired t-test was used and the data are presented as mean ± SEM.
Immunohistochemistry
Mouse brains were fixed by perfusion with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde in PBS 1 month after virus injection. The brains were post-fixed in the same fixative overnight for preparing paraffin or freezing sections (30% sucrose in 0.1 M of PBS for 2 days after post-fixation). The coronal paraffin sections were cut at 5 μm thicknesses for beta-amyloid immunohistochemical staining. The sections were then incubated in 0.3% H2O2 at 37℃, followed by incubation for 2 h in blocking buffer (TBS containing 0.3% Triton X-100 and 5% normal horse serum). Next, the sections were incubated overnight with monoclonal anti-beta-amyloid 1–42 antibody (Millipore, 1:1000) in blocking buffer and then incubated for 2 h with biotinylated horse anti-mouse IgG diluted 1:100 in blocking buffer, followed by incubation for 1 h in an avidin–biotin-peroxidase complex solution.
Quantification of Aβ Level and Positive Plaques
Mouse hippocampi were homogenized in tissue lysate buffer (20 mM Tris–HCl (pH 7.4), 1 mM ethylenediaminetetraacetic acid, 1 mM ethylene glycol tetraacetic acid, 250 mM sucrose) supplemented with protease inhibitors (Roche). The tissue (n = 3 per group) homogenates were treated with diethanolamine to extract soluble β-amyloid. β-amyloid was measured using Invitrogen Aβ 40 Mouse ELISA Kit (Cat# KMB3481) according to the manufacturer’s protocol. The statistical significance was calculated using an unpaired t-test and the data are presented as mean ± SEM.
The stained brain sections were analyzed and photographed using a Zeiss Axioplan microscope using AxioVision 4.6 software. The distribution and quantity of Aβ-positive plaques was determined in AAV-GFP and AAV-GFP-miR-650 virus injected mice (n = 3 per group), and 5 sections per dentate gyrus were analyzed for plaque load. Plaque loads were quantified using ImageJ software and recorded total numbers. The numbers of plaque load were averaged from 3 mice in each group and compared. The statistical significance was calculated using an unpaired t-test and the data are presented as mean ± SEM.
Immunofluorescence Staining
Mice were perfused with PBS followed by 4% paraformaldehyde 1 month after virus injection and brains were removed and dehydrated in 30% sucrose. 40 µm coronal brain sections were collected on a Leica cryostat. The immunofluorescence staining was performed as described before [40]. The primary anti-mouse MAP2 antibody (Sigma, 1:500) and the secondary antibody goat anti-mouse 568 (Invitrogen, 1:500) were applied sequentially. The slides were studied on a Zeiss microscope.
Bioinformatic Analysis
A online program TargetScan was used to predict targets of miR-650 by searching conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA [41] (https://www.targetscan.org/vert_80/). From all potential targets, we selected key genes that involved in the pathogenesis of AD for further analysis.
Statistical Analysis
Statistical analysis was performed using one-way ANOVA and unpaired t-test. All data are presented as mean with standard error of mean (mean ± SEM).
Results
Significantly Increased Expression of miR-650 in the Brain of AD Patients
To analyze the expression of miRNAs and reveal the specific miRNA changes in the brain samples of AD patients, a total of 204 miRNAs were analyzed using TaqMan assays (Table 2), and 17 miRNAs were significantly increased in AD samples compared to control samples (p < 0.05) (Fig. 1A, top 17). MiR-650 stood out as the most significantly increased one among 17 miRNAs (p = 0.00236, Table 2).
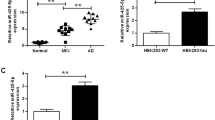
Identification of significantly altered miRNAs in brain from AD patients. A Relative expression levels of top 17 miRNAs from control and Alzheimer’s disease (AD) subjects. For each miRNA, left column represents the control and the right column represents the AD. Unpaired t-test, p-value showed on the top of each group. B–D Relative expression levels of matured miR-650 (B), primary miR-650 (C), and precursor and primary transcript of miR-650 (D) for control and AD subjects. Unpaired t-test, p < 0.01. Control, n = 16; AD, n = 18
Disruption of Production of Mature miR-650 in AD Cases
MiRNAs are generated from long primary miRNAs and processed into precursor miRNAs, and finally formed mature miRNAs (15–18). Deregulated processing of miRNA, as one of the mechanisms to regulate miRNAs expression, has been observed in multiple diseases [17, 20]. We hypothesized that the increased miR-650 expression observed in AD cases might be due to an alteration in miRNA processing. Mature form of miR-650 was generated from the 5-prime strand of its precursor (Supplemental Fig. 1). To test this hypothesis, we analyzed the expression level of primary transcripts, precursor transcripts, and the mature form of miR-650 by quantitative real-time PCR. Surprisingly, we found that in contrast to the increased mature form of miR-650, levels of primary and precursor forms of miR-650 were significantly decreased in AD cases compared to controls (Fig. 1B–D). These results indicate that the processing of primary miR-650 to mature miR-650 are altered in AD patients.
Bioinformatic Prediction of miR-650 Targets Related to AD Pathogenesis
We next performed bioinformatic analyses to identify the potential targets for the increased miRNAs. Utilizing commercially available software programs TargetScan, we found miR-650, which was significantly increased in AD samples, was predicted to bind the 3′ untranslated regions (3′-UTR) of Apolipoprotein E (APOE), Presenilin 1 (PSEN1), and Cyclin-Dependent Kinase 5 (CDK5) (Supplemental Fig. 2).
miR-650 Is Able to Regulate Multiple Genes Implicated in AD
The prominent function of miRNA is to post-transcriptionally regulate the expression target genes [17, 19, 20]. To further study the function of miR-650, we cloned the 3′-UTR of APOE, PSEN1, and CDK5 into a luciferase vector and co-transfected each with miR-650 into HEK293 cells. Target alignment sequence of each 3′UTR to miR-650 is shown in Fig. 2A–C. To avoid non-specific effects, we used miR-572 as a negative control, as it is not predicted to target any of these three genes. The cells were collected 48 h post-transfection, and the luciferase assay was performed. Compared to control, miR-650 was able to robustly decrease luciferase activity (Fig. 2D–F). These results indicate miR-650 can bind to the 3′UTR of the predicted targets APOE, PSEN1, and CDK5.
Expression with miR-650 significantly decreases expression of APOE, PSEN1, and CDK5 in vitro. A–C Predicted target alignment sequence for miR-650 with APOE, PSEN1, and CDK5 3′UTR. Also depicted is mutant 3′UTR vectors with deletion of miR-650 binding site sequence. D–F Expression of miR-650 leads to decreased expression of APOE (D), PSEN1 (E), and CDK5 (F) as assessed by a luciferase reporter. miR-572 was used as a negative control as it is not predicted to target any of these genes. G–I Deletion of miR-650 target sequence abolishes the significant finding of APOE (G), PSEN 1 (H), and CDK5 (I). Values are mean ± SD from three independent experiments; each experiment included triplicate for every co-transfection. One-way ANOVA, p < 0.01, n.s., no significant
To further validate these results, we constructed mutant 3′-UTR-luciferase vectors by deleting the binding sites of miR-650 in the 3′-UTRs of the predicted targets (Fig. 2A–C). Cotransfection and luciferase assays were performed as described above, and, as expected, miR-650 was no longer able to significantly decrease the luciferase activity (Fig. 2G–I). Together, these data indicate that miR-650 is able to specifically regulate the expression of APOE, PSEN1, and CDK5.
Overexpression of miR-650 Downregulates CDK5 and Ameliorates AD Pathogenesis in APP/PSEN1 Transgenic Mice
We next investigated whether overexpression of miR-650 could modulate the expression of the predicted target CDK5 in vivo. To perform this experiment, we used APP/PSEN1 double transgenic mice and measured CDK5 levels. APP/PSEN1 double transgenic mice contain mutant forms of APP and PSEN1 that are associated with early onset AD [42]. Virus injections were performed bilaterally on the hippocampus of each mouse (n = 5): injection was administered at the left lateral DG for AAV-GFP and the right lateral DG for AAV-miR-650 (Fig. 3A). One month after virus grafting, the mice were sacrificed, and hippocampi were dissected out. To confirm the efficiency of virus injection and infection, we first performed western blot to analyze the expression of GFP. We found both AAV-GFP and AAV-GFP-miR-650 grafted samples displayed equal levels of expression of GFP, confirming the efficiency of the injection and virus infection in vivo (Supplemental Fig. 3); importantly, miR-650 expression was only seen with the AAV-miR-650 samples (Fig. 3B). These samples were also used to measure CDK5 levels, and we found that CDK5 protein (Fig. 3C–D) and transcript levels (Fig. 3E) was significantly decreased by over-expressing miR-650. These data indicated that miR-650 can regulate CDK5 protein levels in vivo. Taken together, these data further confirmed CDK5 is a bona fide target of miR-650 in vivo.
Overexpression of miR-650 decreases CDK5 levels in vivo. A The dentate gyrus (DG) of APP-PSEN1 mice was injected bilaterally with adeno-associated virus (AAV) vectors: AAV-GFP control virus was injected in the left DG and AAV-miR-650 virus in the right DG (n = 3). B Expression of miR-650 was only seen in the hippocampus sections that were injected with AAV-miR-650 (unpaired t-test, p < 0.01). C–D Western blot analysis shows a significant decrease in CDK5 levels on hippocampus samples injected with AAV-miR-650 (unpaired t-test, p < 0.01). E Transcript levels of CDK5 were also significantly decreased in the samples injected with AAV-miR-650 (unpaired t-test, p < 0.01)
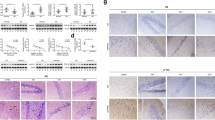
To determine if reduced CDK5 induced by miR-650 overexpression has neuronal effects, we performed immunostaining of neuronal marker microtubule-associated protein 2 (MAP2) with sections from virus injected APP/PSEN1 mice [43]. First, the staining revealed that in the presence of overexpressed miR-650 the number of plaques and Aβ levels were significantly reduced (Fig. 4A–D). Furthermore, the overexpression of miR-650 also significantly increased MAP2 positive cells (neuronal cells) in the dentate gyrus of APP/PSEN1 mice, compared to GFP virus injection (Fig. 4E–K). These results suggest miR-650 has the ability to ameliorate neuronal deficits in APP mice.
Overexpression of miR-650 ameliorates AD pathogenesis in APP/PSEN1 transgenic mice. A–B Immunohistochemistry of beta-amyloid for hippocampal sections from AAV-GFP control mice (A) and AAV-miR-650 mice (B). C AAV-miR-650 brain sections show significantly fewer number of plaques. D AAV-miR-650 mice show significantly lower amyloid-β (Aβ) levels (n = 3, unpaired t-test, p < 0.05). E–J Immunostaining of hippocampal sections for AAV-GFP control mice (E–G) and AAV-miR-650 mice (H–J). Neurons are indicated by red staining with MAP2 antibody. Arrow indicates increased neuronal staining in the presence of miR-650 overexpression. K AAV-miR-650 mice show significantly increased number of neuronal cells (n = 3, unpaired t-test, p < 0.05)
Discussion
In this study, we described a novel miR-650-CDK5 regulatory axis that is involved in the physiopathology of AD. Significantly increased miR-650 was found in sporadic AD patients. Furthermore, in vitro assay demonstrated that APOE, PSEN1, and CDK5 are all specifically targeted by miR-650. Importantly, CDK5 expression was shown to be regulated by miR-650 in vivo using a mouse model for early onset AD: overexpression of miR-650 significantly inhibited CDK5 expression and increased the immunostaining intensity of neuronal cells in APP/PSEN1 mice.
Irregular expression of miR-650 has been well documented in multiple types of human cancer. Several targets of miRNA-650 have been identified, including ING4, CDK1, AKT2, EBF3, and other factors [44,45,46,47,48,49,50,51,52,53,54,55]. Nonetheless, the importance of miR-650 in AD pathogenesis has not been defined. In our study, we observed a significant increase of miR-650 in AD patients. Predicted by bioinformatic analysis that CDK5 is one of the targets of miR-650, the direct interaction of miR-650 with CDK5 3′-UTR has been validated by in vitro luciferase functional studies using normal and mutated CDK5 3′-UTR structures. Furthermore, an inverse relationship between the expression of miR-650 and CDK5 was demonstrated by in vivo overexpression of miR-650 by adeno-associated virus leading to reduced CDK5 expression. Thus, our data suggested an important molecular link between miR-650 and CDK5 in AD mouse model.
CDK5 is an atypical proline-guided serine/threonine kinase that phosphorylates various substrate and exerts multiple regulatory roles in neuronal cells, as well as other cell types [56,57,58]. CDK5 is required for normal brain development, as evidenced by Cdk5-null mice not being viable past postnatal day zero [59]. The CDK5/p25 complex hyperphosphorylates Tau and leads to irreversible cytoskeletal disruption of neuronal cells and induced neuronal cell death [60]. Previous studies have demonstrated that CDK5 activity is increased in several different neurodegenerative diseases including AD, animal models of amyotrophic lateral sclerosis (ALS) and Niemann Pick type C (NPC) disease [61,62,63]. By manipulating CDK5 expression via overexpression of miR-650 in an AD mouse model, we showed that reduced plaques and Aβ presence in the brain. Thus, the miR-650-CDK5 regulatory axis defined in our study has important implications in AD pathogenesis.
Recent progress in the understanding of AD pathogenesis indicated that dysregulation of miRNA biogenesis was involved in neurodegeneration. On this regard, new methodological advances of AD therapy have developed short and synthetic antisense oligonucleotides (ASOs), that recognize target mRNA for posttranscriptional regulation to correct protein expression errors. The miRNA based ASOs design provided a possibility for synchronous regulation of protein expression and related signaling pathways [64, 65]. Thus, our finding about miR-650 involved in AD pathogenesis provides a potential target to develop ASOs therapeutics for AD.
In summary, our present results showed that miR-650 can regulate CDK5 both in vitro and in vivo. Over-expression of miR-650 can also significantly ameliorate neuronal deficits. These results suggest miR-650 is actively involved in neurodegeneration through modulating CDK5.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297(5580):353–356. https://doi.org/10.1126/science.1072994
Tanzi RE, Bertram L (2005) Twenty years of the Alzheimer’s disease amyloid hypothesis: a genetic perspective. Cell 120(4):545–555. https://doi.org/10.1016/j.cell.2005.02.008
Selkoe DJ, Hardy J (2016) The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med 8(6):595–608. https://doi.org/10.15252/emmm.201606210
De-Paula VJ, Radanovic M, Diniz BS, Forlenza OV (2012) Alzheimer’s disease. Subcell Biochem 65:329–352. https://doi.org/10.1007/978-94-007-5416-4_14
Nizynski B, Dzwolak W, Nieznanski K (2017) Amyloidogenesis of Tau protein. Protein Sci Publ Protein Soc 26(11):2126–2150. https://doi.org/10.1002/pro.3275
Cohen P (2001) The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem 268(19):5001–5010. https://doi.org/10.1046/j.0014-2956.2001.02473.x
Pawson T, Scott JD (2005) Protein phosphorylation in signaling–50 years and counting. Trends Biochem Sci 30(6):286–290. https://doi.org/10.1016/j.tibs.2005.04.013
Tenreiro S, Eckermann K, Outeiro TF (2014) Protein phosphorylation in neurodegeneration: friend or foe? Front Mol Neurosci 7:42. https://doi.org/10.3389/fnmol.2014.00042
Henriques AG, Muller T, Oliveira JM, Cova M, da Cruz ESCB, da Cruz ESOA (2016) Altered protein phosphorylation as a resource for potential AD biomarkers. Sci Rep 6:30319. https://doi.org/10.1038/srep30319
Llorens-Martin M, Jurado J, Hernandez F, Avila J (2014) GSK-3beta, a pivotal kinase in Alzheimer disease. Front Mol Neurosci 7:46. https://doi.org/10.3389/fnmol.2014.00046
Lukasik P, Zaluski M, Gutowska I (2021) Cyclin-dependent kinases (CDK) and their role in diseases development-review. Int J Mol Sci 22(6). https://doi.org/10.3390/ijms22062935
Tapia-Rojas C, Cabezas-Opazo F, Deaton CA, Vergara EH, Johnson GVW, Quintanilla RA (2019) It’s all about tau. Prog Neurobiol 175:54–76. https://doi.org/10.1016/j.pneurobio.2018.12.005
Iqbal K, Liu F, Gong CX (2016) Tau and neurodegenerative disease: the story so far. Nat Rev Neurol 12(1):15–27. https://doi.org/10.1038/nrneurol.2015.225
Hamdane M, Bretteville A, Sambo AV, Schindowski K, Begard S, Delacourte A, Bertrand P, Buee L (2005) p25/Cdk5-mediated retinoblastoma phosphorylation is an early event in neuronal cell death. J Cell Sci 118(Pt 6):1291–1298. https://doi.org/10.1242/jcs.01724
Jiang XY, Chen TK, Zhou JT, He SY, Yang HY, Chen Y, Qu W, Feng F et al (2018) Dual GSK-3beta/AChE inhibitors as a new strategy for multitargeting anti-Alzheimer’s disease drug discovery. ACS Med Chem Lett 9(3):171–176. https://doi.org/10.1021/acsmedchemlett.7b00463
Jadhav S, Avila J, Scholl M, Kovacs GG, Kovari E, Skrabana R, Evans LD, Kontsekova E et al (2019) A walk through tau therapeutic strategies. Acta Neuropathol Commun 7(1):22. https://doi.org/10.1186/s40478-019-0664-z
Michlewski G, Caceres JF (2019) Post-transcriptional control of miRNA biogenesis. RNA 25(1):1–16. https://doi.org/10.1261/rna.068692.118
Wang M, Qin L, Tang B (2019) MicroRNAs in Alzheimer’s disease. Front Genet 10:153. https://doi.org/10.3389/fgene.2019.00153
Wei W, Wang ZY, Ma LN, Zhang TT, Cao Y, Li H (2020) MicroRNAs in Alzheimer’s disease: function and potential applications as diagnostic biomarkers. Front Mol Neurosci 13:160. https://doi.org/10.3389/fnmol.2020.00160
Lekka E, Hall J (2018) Noncoding RNAs in disease. FEBS Lett 592(17):2884–2900. https://doi.org/10.1002/1873-3468.13182
Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH (2006) Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci 29:77–103
Li X, Jin P (2010) Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci 11(5):329–338. https://doi.org/10.1038/nrn2739
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. https://doi.org/10.1016/j.cell.2009.01.002
Bushati N, Cohen SM (2008) MicroRNAs in neurodegeneration. Curr Opin Neurobiol 18(3):292–296
Eacker SM, Dawson TM, Dawson VL (2009) Understanding microRNAs in neurodegeneration. Nat Rev Neurosci 10(12):837–841. https://doi.org/10.1038/nrn2726
Vishnoi A, Rani S (2017) MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol 1509:1–10. https://doi.org/10.1007/978-1-4939-6524-3_1
Juzwik CA, Drake SS, Zhang Y, Paradis-Isler N, Sylvester A, Amar-Zifkin A, Douglas C, Morquette B et al (2019) microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurobiol 182:101664. https://doi.org/10.1016/j.pneurobio.2019.101664
Bettens K, Brouwers N, Engelborghs S, Van Miegroet H, De Deyn PP, Theuns J, Sleegers K, Van Broeckhoven C (2009) APP and BACE1 miRNA genetic variability has no major role in risk for Alzheimer disease. Hum Mutat 30(8):1207–1213. https://doi.org/10.1002/humu.21027
Boissonneault V, Plante I, Rivest S, Provost P (2009) MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J Biol Chem 284(4):1971–1981. https://doi.org/10.1074/jbc.M807530200
Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A et al (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A 105(17):6415–6420. https://doi.org/10.1073/pnas.0710263105
Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT (2008) The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci 28(5):1213–1223
Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y et al (2009) miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull 80(4–5):268–273. https://doi.org/10.1016/j.brainresbull.2009.08.006
Maciotta S, Meregalli M, Torrente Y (2013) The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci 7:265. https://doi.org/10.3389/fncel.2013.00265
Su L, Li R, Zhang Z, Liu J, Du J, Wei H (2022) Identification of altered exosomal microRNAs and mRNAs in Alzheimer’s disease. Ageing Res Rev 73:101497. https://doi.org/10.1016/j.arr.2021.101497
Goh SY, Chao YX, Dheen ST, Tan EK, Tay SS (2019) Role of MicroRNAs in Parkinson’s disease. Int J Mol Sci 20(22). https://doi.org/10.3390/ijms20225649
McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH, Klunk WE, Koroshetz WJ et al (2011) The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement J Alzheimers Assoc 7(3):263–269. https://doi.org/10.1016/j.jalz.2011.03.005
Jardanhazi-Kurutz D, Kummer MP, Terwel D, Vogel K, Dyrks T, Thiele A, Heneka MT (2010) Induced LC degeneration in APP/PS1 transgenic mice accelerates early cerebral amyloidosis and cognitive deficits. Neurochem Int 57(4):375–382. https://doi.org/10.1016/j.neuint.2010.02.001
Kummer MP, Hammerschmidt T, Martinez A, Terwel D, Eichele G, Witten A, Figura S, Stoll M et al (2014) Ear2 deletion causes early memory and learning deficits in APP/PS1 mice. J Neurosci Off J Soc Neurosci 34(26):8845–8854. https://doi.org/10.1523/JNEUROSCI.4027-13.2014
Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W et al (2010) Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol 189(1):127–141. https://doi.org/10.1083/jcb.200908151
Li X, Barkho BZ, Luo Y, Smrt RD, Santistevan NJ, Liu C, Kuwabara T, Gage FH et al (2008) Epigenetic regulation of the stem cell mitogen Fgf-2 by Mbd1 in adult neural stem/progenitor cells. J Biol Chem 283(41):27644–27652. https://doi.org/10.1074/jbc.M804899200
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20. https://doi.org/10.1016/j.cell.2004.12.035
Schmitz C, Rutten BP, Pielen A, Schafer S, Wirths O, Tremp G, Czech C, Blanchard V et al (2004) Hippocampal neuron loss exceeds amyloid plaque load in a transgenic mouse model of Alzheimer’s disease. Am J Pathol 164(4):1495–1502. https://doi.org/10.1016/S0002-9440(10)63235-X
Izant JG, McIntosh JR (1980) Microtubule-associated proteins: a monoclonal antibody to MAP2 binds to differentiated neurons. Proc Natl Acad Sci U S A 77(8):4741–4745. https://doi.org/10.1073/pnas.77.8.4741
Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu Z, Zhang M (2010) MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun 395(2):275–280. https://doi.org/10.1016/j.bbrc.2010.04.005
Chien WW, Domenech C, Catallo R, Kaddar T, Magaud JP, Salles G, Ffrench M (2011) Cyclin-dependent kinase 1 expression is inhibited by p16(INK4a) at the post-transcriptional level through the microRNA pathway. Oncogene 30(16):1880–1891. https://doi.org/10.1038/onc.2010.570
Huang JY, Cui SY, Chen YT, Song HZ, Huang GC, Feng B, Sun M, De W et al (2013) MicroRNA-650 was a prognostic factor in human lung adenocarcinoma and confers the docetaxel chemoresistance of lung adenocarcinoma cells via regulating Bcl-2/Bax expression. PloS one 8(8):e72615. https://doi.org/10.1371/journal.pone.0072615
Zhou C, Cui F, Li J, Wang D, Wei Y, Wu Y, Wang J, Zhu H et al (2017) MiR-650 represses high-risk non-metastatic colorectal cancer progression via inhibition of AKT2/GSK3beta/E-cadherin pathway. Oncotarget 8(30):49534–49547. https://doi.org/10.18632/oncotarget.17743
Ye Y, Zhuang J, Wang G, He S, Ni J, Xia W, Wang J (2017) microRNA-605 promotes cell proliferation, migration and invasion in non-small cell lung cancer by directly targeting LATS2. Exp Ther Med 14(1):867–873. https://doi.org/10.3892/etm.2017.4538
Mraz M, Dolezalova D, Plevova K, Stano Kozubik K, Mayerova V, Cerna K, Musilova K, Tichy B et al (2012) MicroRNA-650 expression is influenced by immunoglobulin gene rearrangement and affects the biology of chronic lymphocytic leukemia. Blood 119(9):2110–2113. https://doi.org/10.1182/blood-2011-11-394874
Pichulik T, Khatamzas E, Liu X, Brain O, Delmiro Garcia M, Leslie A, Danis B, Mayer A et al (2016) Pattern recognition receptor mediated downregulation of microRNA-650 fine-tunes MxA expression in dendritic cells infected with influenza A virus. Eur J Immunol 46(1):167–177. https://doi.org/10.1002/eji.201444970
Xu X, Zhu X, Wang C, Li Y, Fan C, Kao X (2019) microRNA-650 promotes inflammation induced apoptosis of intestinal epithelioid cells by targeting NLRP6. Biochem Biophys Res Commun 517(4):551–556. https://doi.org/10.1016/j.bbrc.2019.06.077
You Q, Li H, Liu Y, Xu Y, Miao S, Yao G, Xue Y, Geng J et al (2018) MicroRNA-650 targets inhibitor of growth 4 to promote colorectal cancer progression via mitogen activated protein kinase signaling. Oncol Lett 16(2):2326–2334. https://doi.org/10.3892/ol.2018.8910
Jin P, Chen H, Xie J, Zhou C, Zhu X (2017) Essential role of microRNA-650 in the regulation of B-cell CLL/lymphoma 11B gene expression following transplantation: A novel mechanism behind the acute rejection of renal allografts. Int J Mol Med 40(6):1840–1850. https://doi.org/10.3892/ijmm.2017.3194
Tang X, Ding Y, Wang X, Wang X, Zhao L, Bi H (2019) miR-650 promotes non-small cell lung cancer cell proliferation and invasion by targeting ING4 through Wnt-1/beta-catenin pathway. Oncol Lett 18(5):4621–4628. https://doi.org/10.3892/ol.2019.10805
Jin S, Li X, Dai Y, Li C, Wang D (2020) NF-kappaB-mediated miR-650 plays oncogenic roles and activates AKT/ERK/NF-kappaB pathways by targeting RERG in glioma cells. Cell Oncol 43(6):1035–1048. https://doi.org/10.1007/s13402-020-00533-5
Liu SL, Wang C, Jiang T, Tan L, Xing A, Yu JT (2016) The role of Cdk5 in Alzheimer’s disease. Mol Neurobiol 53(7):4328–4342. https://doi.org/10.1007/s12035-015-9369-x
Shah K, Lahiri DK (2014) Cdk5 activity in the brain - multiple paths of regulation. J Cell Sci 127(Pt 11):2391–2400. https://doi.org/10.1242/jcs.147553
Sharma S, Sicinski P (2020) A kinase of many talents: non-neuronal functions of CDK5 in development and disease. Open Biol 10(1):190287. https://doi.org/10.1098/rsob.190287
Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna PHC, Brady RO, Martin LJ, Kulkarni AB (1996) Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci U S A 93(20):11173–11178
Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH (1999) Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature 402(6762):615–622. https://doi.org/10.1038/45159
Bu B, Li J, Davies P, Vincent I (2002) Deregulation of cdk5, hyperphosphorylation, and cytoskeletal pathology in the Niemann-Pick type C murine model. J Neurosci 22(15):6515–6525
Lee KY, Clark AW, Rosales JL, Chapman K, Fung T, Johnston RN (1999) Elevated neuronal Cdc2-like kinase activity in the Alzheimer disease brain. Neurosci Res 34(1):21–29
Nguyen MD, Lariviere RC, Julien JP (2001) Deregulation of Cdk5 in a mouse model of ALS: toxicity alleviated by perikaryal neurofilament inclusions. Neuron 30(1):135–147
Grabowska-Pyrzewicz W, Want A, Leszek J, Wojda U (2021) Antisense oligonucleotides for Alzheimer’s disease therapy: from the mRNA to miRNA paradigm. EBioMedicine 74:103691. https://doi.org/10.1016/j.ebiom.2021.103691
Roy B, Lee E, Li T, Rampersaud M (2022) Role of miRNAs in neurodegeneration: from disease cause to tools of biomarker discovery and therapeutics. Genes 13(3). https://doi.org/10.3390/genes13030425
Acknowledgements
We would like to thank Dr. Junwei Zeng for critical reading and editing of the manuscript.
Funding
L.L is supported by grants from Natural Science Foundation of Guangdong Province (2019A1515011416, 2022A1515010689), Key Field Research and Development Program of Guangdong province (2018B030337001), Guangzhou Key Research Program on Brain Science (202007030008), and Guangdong Key Laboratory of nonhuman primate models of brain diseases. X.L. is supported by the National Key Basic Research Program of China (2014CB943001) and National Natural Science Foundation of China (31371309).
Author information
Authors and Affiliations
Contributions
L.L, P.J, and XK.L conceived the project. L.L, XJ.C XD.L, YJ.L, M.G, A.L, XL. H, Y.L, and XK. L performed the experiments and statistics. L.L, P.J, and XK.L interpreted results and drafted the manuscript. All authors edited and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
Brain tissues were obtained from archival human brain specimens of the Emory ADRC Neuropathology Core, using appropriate IRB protocols. Mice were kept at the animal facility at Emory University. All animal experiments were performed in accordance with protocols approved in advance by Emory University (IACUC).
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
The authors affirm that human research participants provided informed consent for publication.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, L., Liu, X., Cheng, X. et al. MicroRNA-650 Regulates the Pathogenesis of Alzheimer’s Disease Through Targeting Cyclin-Dependent Kinase 5. Mol Neurobiol 60, 2426–2441 (2023). https://doi.org/10.1007/s12035-023-03224-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03224-y