Abstract
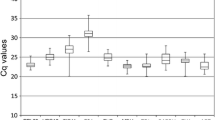
The selection of appropriate reference genes is one of the most important steps to obtain reliable results for normalizing quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) of MADS-box gene in Phalaenopsis. In this study, we cloned 12 candidate reference genes including 18S ribosomal RNA (18S), elongation factor 1 alpha (EF1α), cytoskeletal structural protein actin (ACT1, ACT2, ACT3, ACT4, ACT5), ubiquitin protein (UBQ1 and UBQ2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the cytoskeletal structural proteins α-tubulin (TUA) and β-tubulin (TUB) in Phalaenopsis and evaluated their expression reliability. The expression of these candidate reference genes was analyzed using geNorm and normFinder software packages; the results showed that ACT2 and ACT4 were the highest stability reference genes for all experiment sets based on normFinder, followed by ACT1 or ACT3, while ACT3 and ACT4 were the highest stability reference genes for most experiment sets based on geNorm, then TUB or others. Taken together, Actin genes were the higher stability reference genes for all tissues at total developmental stages, and similar results came from analysis by normFinder. According to geNorm analysis, ACT3 and ACT4 were the most stable reference genes for all tissues tested and tissues at reproductive stages; TUB and ACT5 or ACT4 were the most stable reference genes for vegetative tissues or roots. The most stable reference genes for all vegetative tissues and only leaves were ACT4 and ACT5, ACT2 and ACT3, respectively; ACT1 and ACT3 were the most stable genes and sufficient for reliable normalization of flower tissues. While EF1α, UBQ1, UBQ2, and GAPDH were found to be unsuitable as a reference gene in our analysis for flower tissues, total tissues, and reproductive stages; UBQ2 and 18S were identified as the least stable reference genes for vegetative tissues at different stages, different tissues at vegetative stages; TUA and 18S were the least reliable reference genes for the samples from roots at all developmental stages. This is the first systematic report on the selection of reference genes in Phalaenopsis, and these data will facilitate future work on gene expression in orchid.




Similar content being viewed by others
References
Rudall, P. J., & Bateman, R. M. (2002). Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biological Reviews, 77, 403–441. doi:10.1017/s1464793102005936.
Mondragón-Palomino, M., & Theissen, G. (2009). Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Annals of Botany, 104, 583–594. doi:10.1093/aob/mcn258.
Tsai, W. C., & Chen, H. H. (2006). The orchid MADS-Box genes controlling floral morphogenesis. Scientific World Journal, 6, 1933–1944. doi:10.1100/tswde.2006.321.
Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., et al. (2006). The real-time polymerase chain reaction. Molecular Aspects of Medicine, 27, 95–125. doi:10.1016/j.mam.2005.12.007.
Bustin, S. A., Benes, V., Nolan, T., & Pfaffl, M. W. (2005). Quantitative real-time RT-PCR—a perspective. Journal of Molecular Endocrinology, 34, 597–601. doi:10.1677/jme.1.01755.
Dheda, K., Huggett, J. F., Chang, J. S., Kim, L. U., Bustin, S. A., Johnson, M. A., et al. (2005). The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Analytical Biochemistry, 344, 141–143. doi:10.1016/j.ab.2005.05.022.
Huggett, J., Dheda, K., Bustin, S., & Zumla, A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes and Immunity, 6, 279–284. doi:10.1038/sj.gene.6364190.
Radonić, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., & Nitsche, A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochemical and Biophysical Research Communications, 313, 856–862. doi:10.1016/j.bbrc.2003.11.177.
Maroufi, A., Van Bockstaele, E., De Loose, M., Maroufi, A., Bockstaele, E. V., & Loose, M. D. (2010). Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Molecular Biology, 11, 15–27. doi:10.1186/1471-2199-11-15.
Hu, R., Fan, C., Li, H., Zhang, Q., & Fu, Y. F. (2009). Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Molecular Biology, 10, 93–104. doi:10.1186/1471-2199-10-93.
Garg, R., Sahoo, A., Tyagi, A. K., & Jain, M. (2010). Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochemical and Biophysical Research Communications, 396, 283–288. doi:10.1016/j.bbrc.2010.04.079.
Gu, C., Chen, S., Liu, Z., Shan, H., Luo, H., Guan, Z., et al. (2011). Reference gene selection for quantitative real-time PCR in Chrysanthemum subjected to biotic and abiotic stress. Molecular Biotechnology, 49, 192–197. doi:10.1007/s12033-011-9394-6.
Schmittgen, T. D., & Zakrajsek, B. A. (2000). Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. Journal of Biochemical and Biophysical Methods, 46, 69–81. doi:10.1016/S0165-022X(00)00129-9.
Jain, M., Nijhawan, A., Tyagi, A. K., & Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications, 345, 646–651. doi:10.1016/j.bbrc.2006.04.140.
Jian, B., Liu, B., Bi, Y., Hou, W., Wu, C., & Han, T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Molecular Biology, 9, 59. doi:10.1186/1471-2199-9-59.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3, 1–11.
Wan, H., Zhao, Z., Qian, C., Sui, Y., Malik, A. A., & Chen, J. (2010). Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Analytical Biochemistry, 399, 257–261. doi:10.1016/j.ab.2009.12.008.
Qi, J., Yu, S., Zhang, F., Shen, X., Zhao, X., Yu, Y., et al. (2010). Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese abbage (Brassica rapa L. ssp. pekinensis). Plant Mol Biol Rep, 28, 597–604. doi:10.1007/s11105.-010-0185-1.
Yan, J., Yuan, F., Long, G., Qin, L., & Deng, Z. (2012). Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Molecular Biology Reports, 39, 1831–1838. doi:10.1007/s11033-011-0925-9.
Chen, L., Zhong, H. Y., Kuang, J. F., Li, J. G., Lu, W. J., & Chen, J. Y. (2011). Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta, 234, 377–390. doi:10.1007/s00425-011-1410-3.
Podevin, N., Krauss, A., Henry, I., Swennen, R., & Remy, S. (2012). Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol Breeding., 30, 1237–1252. doi:10.1007/s11032-012-9711-1.
Long, X. Y., Wang, J. R., Ouellet, T., Rocheleau, H., Wei, Y. M., Pu, Z. E., et al. (2010). Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Molecular Biology, 74, 307–311. doi:10.1007/s11103-010-9666-8.
Chen, Y. Y., Lee, P. F., Hsiao, Y. Y., Wu, W. L., Pan, Z. J., Lee, Y. I., et al. (2012). C-and D-class MADS-Box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant Cell Physiology, 53, 1053–1067. doi:10.1093/pcp/pcs048.
Song, I. J., Fukuda, T., ITO, T., KO, S. T., Yokoyama, J., Ichikawa, H., et al. (2011). Expression analysis of an APETALA1/FRUITFULL-like gene in Phalaenopsis sp. ‘Hatsuyuki’ (Orchidaceae). Horticulture Environmental Biotechnology, 52, 183–195. doi:10.1007/s13580-011-0199-0.
Chen, D., Guo, B., Hexige, S., Zhang, T., Shen, D., & Ming, F. (2007). SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta, 226, 369–380. doi:10.1007/s00425-007-0488-0.
Andersen, C. L., Jensen, J. L., & Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250.
Heuer, S., Hansen, S., Bantin, J., Brettschneider, R., Kranz, E., Lörz, H., et al. (2001). The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiology, 127, 33–45.
Chen, M. K., Lin, I. C., & Yang, C. H. (2008). Functional analysis of three lily (Lilium longiflorum) APETALA1-like MADS box genes in regulating floral transition and formation. Plant Cell Physiology, 49, 704–717. doi:10.1093/pcp/pcn046.
Guénin, S., Mauriat, M., Pelloux, J., Van Wuytswinkel, O., Bellini, C., & Gutierrez, L. (2009). Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of reference. Journal of Experimental Botany, 60, 487–493. doi:10.1093/jxb/ern305.
Nicot, N., Hausman, J. F., Hoffmann, L., & Evers, D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany, 56, 2907–2914. doi:10.1093/jxb/eri285.
Lovdal, T., & Lillo, C. (2009). Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Analytical Biochemistry, 387, 238–242. doi:10.1016/j.ab.2009.01.024.
Tong, Z., Gao, Z., Wang, F., Zhou, J., & Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Molecular Biology, 10, 71–82. doi:10.1186/1471-2199-10-71.
Brunner, A. M., Yakovlev, I. A., & Strauss, S. H. (2004). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology, 4, 14. doi:10.1186/1471-2229-4-14.
González-Verdejo, C. I., Die, J. V., Nadal, S., Jiménez-Marín, A., Moreno, M. T., & Román, B. (2008). Selection of housekeeping genes for normalization by real-time RT-PCR: analysis of Or-MYB1 gene expression in Orobanche ramosa development. Analytical Biochemistry, 379, 176–181. doi:10.1016/j.ab.2008.05.003.
Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F., & Park, C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biology, 8, 112. doi:10.1186/1471-2229-8-112.
Iskandar, H. M., Simpson, R. S., Casu, R. E., Bonnett, G. D., Maclean, D. J., & Manners, J. M. (2004). Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Mol Biol Rep., 22, 325–337. doi:10.1007/BF02772676.
Singh, R., & Green, M. R. (1993). Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science, 259, 365–368. doi:10.1126/science.8420004.
Ishitani, R., Sunaga, K., Hirano, A., Saunders, P., Katsube, N., & Chuang, D. M. (1999). Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. Journal of Neurochemistry, 66, 928–935. doi:10.1046/j.1471-4159.1996.66030928.x.
Li, H., Qin, Y., Xiao, X., & Tang, C. (2011). Screening of valid reference genes for real-time RT-PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Science, 181, 132–139. doi:10.1016/j.plantsci.2011.04.014.
Wasteneys, G. O. (2004). Progress in understanding the role of microtubules in plant cells. Current Opinion in Plant Biology, 7, 651–660. doi:10.1016/j.pbi.2004.09.008.
Yoshikawa, M., Yang, G., Kawaguchi, K., & Komatsu, S. (2003). Expression analyses of beta-tubulin isotype genes in rice. Plant Cell Physiology, 44, 1202–1207. doi:10.1093/pcp/pcg150.
Oakley, R. V., Wang, Y. S., Ramakrishna, W., Harding, S. A., & Tsai, C. J. (2007). Differential expansion and expression of alpha- and beta-tubulin gene families in Populus. Plant Physiology, 145, 961–973. doi:10.1104/pp. 107.107086.
Li, X. S., Yang, H. L., Zhang, D. Y., Zhang, Y. M., & Wood, A. J. (2012). Reference gene selection in the desert plant Eremosparton songoricum. International Journal of Molecular Sciences, 13, 6944–6963. doi:10.3390/ijms13066944.
Marum, L., Miguel, A., Ricardo, C. P., & Miguel, C. (2012). Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PloS One, 7, e35113. doi:10.1371/journal.pone.0035113.
Yang, Y., Hou, S., Cui, G., Chen, S., Wei, J., & Huang, L. (2010). Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Molecular Biology Reports, 37, 507–513. doi:10.1007/s11033-009-9703-3.
Acknowledgments
We thank the orchid center of engineering technology in Zhengzhou for plant materials. This work was supported by the key technology project of Henan Province (092102110128), Zhengzhou natural science project (112PPTGY250-3), and the subject of Zhengzhou Normal University (2012081).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1
(DOC 29 kb)
Supplementary material 2
(DOC 97 kb)
Supplementary material 3
(DOC 5225 kb)
Rights and permissions
About this article
Cite this article
Yuan, XY., Jiang, SH., Wang, MF. et al. Evaluation of Internal Control for Gene Expression in Phalaenopsis by Quantitative Real-Time PCR. Appl Biochem Biotechnol 173, 1431–1445 (2014). https://doi.org/10.1007/s12010-014-0951-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-0951-x