Abstract
Purpose
The role of dominant bacterial groups in the plant rhizosphere, e.g., those belonging to the phyla Acidobacteria and Verrucomicrobia, has, so far, not been elucidated, and this is mainly due to the lack of culturable representatives. This study aimed to isolate hitherto-uncultured bacteria from the potato rhizosphere by a combination of cultivation approaches.
Materials and methods
An agar medium low in carbon availability (oligotrophic agar medium) and either amended with potato root exudates or catalase or left unamended was used with the aim to improve the culturability of bacteria from the potato rhizosphere. The colony forming unit numbers based on colonies and microcolonies were compared with microscopically determined fluorescence-stained cell numbers. Taxonomical diversity of the colonies was compared with that of library clones made from rhizosphere DNA, on the basis of 16S rRNA gene comparisons.
Results and discussion
The oligotrophic media amended or not with catalase or rhizosphere extract recovered up to 33.6% of the total bacterial numbers, at least seven times more than the recovery observed on R2A. Four hitherto-uncultured Verrucomicrobia subdivision 1 representatives were recovered on agar, but representatives of this group were not found in the clone library.
Conclusions
The use of oligotrophic medium and its modifications enabled the growth of colony numbers, exceeding those on classical agar media. Also, it led to the isolation of hitherto-uncultured bacteria from the potato rhizosphere. Further improvement in cultivation will certainly result in the recovery of other as-yet-unexplored bacteria from the rhizosphere, making these groups accessible for further investigation, e.g., with respect to their possible interactions with plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The rhizosphere is a complex environment in which many different microbial species coexist. Some species interact intimately with plant roots, thereby supporting important processes for plant growth, like nutrient cycling and acquisition, disease suppression, and growth stimulation (Curl and Truelove 1986). The microbial community structure in the potato rhizosphere was shown to be influenced by different biotic and abiotic factors like plant growth stage, plant genotype, and local environmental conditions (Rasche et al. 2006; Van Overbeek and van Elsas 2008). The high microbial diversity in the rhizosphere makes it difficult to pinpoint and eventually explore those functions that are important for in situ stimulation of plant health. Exploration of these functions is further complicated because the vast majority of bacterial species is not able to grow on standard agar media (Stevenson et al. 2004). In fact, the taxonomical status of soil isolates obtained by plating on such media will often hardly reflect the composition of the extant bacterial community because the isolation method used only accesses a small subset of this community (Eilers et al. 2000; Felske et al. 1999; Stevenson et al. 2004).
Different approaches have been used to access the microbiota of natural systems, aiming to enhance the culturability of cells extracted from such habitats (Barer and Harwood 1999; Bogosian and Bourneuf 2001; Bruns et al. 2003; Guan and Kamino 2001; Ingham et al. 2007; Janssen et al. 2002; Kamagata and Tamaki 2005; Stevenson et al. 2004; Ueda et al. 2008; Van Overbeek et al. 2004). Such approaches were, for instance, a decrease in nutrient concentrations by the use of dilute media and long incubation periods (Janssen et al. 2002), addition of bacterial signaling molecules (Bruns et al. 2003; Guan and Kamino 2001), replacement of agar by gellan gum as the solidifying agent (Kamagata and Tamaki 2005), and modification of the gas composition during incubation (Stevenson et al. 2004; Ueda et al. 2008). Compounds that reduce the stress imposed by the culture medium can improve culturability, as it was shown that addition of anti-oxidative stress compounds like pyruvate and catalase can lead to a higher recovery of cells from soil and water systems (Barer and Harwood 1999; Bogosian and Bourneuf 2001; Van Overbeek et al. 2004). Also, mimicking environmental nutrient levels rather than using concentrations that are typically used in bacterial growth media can result in the growth of enhanced cell numbers (Hamaki et al. 2005; Kaeberlein et al. 2002). Thus, different hitherto-undescribed species were isolated from soil on low-nutrient media to which soil extract had been added (Hamaki et al. 2005). Clearly, to sensibly enhance bacterial culturability, it is important to know which parameters restrict or prevent growth of bacteria when these are taken out of their natural habitat. Two factors that affect the bacterial response upon transfer from the environment to culture media are exposure to oxidative stress and sudden upshifts in nutrient composition and availability (Barer and Harwood 1999). On low-nutrient media, soil isolates belonging to the unexplored Verrucomicrobia and Acidobacteria have been obtained (Janssen et al. 2002). Members of both groups also prevail in rhizospheres, although culturable representatives are hardly found in this habitat (Buée et al. 2009; Nunes da Rocha et al. 2009). Parameters used to establish the culturability levels of the soil microbiota are (1) the ratio of the colony numbers over the total cells and (2) the diversity of the isolates. A key consideration here is whether the enhanced colony forming unit (CFU) numbers go hand-in-hand with higher diversities of culturable species. The use of parallel and possibly complementary culturing techniques may offer an avenue to achieve a higher diversity of cultured species.
The objective of this study was to evaluate different approaches that may enhance the culturability of bacterial cells from the potato rhizosphere, establishing their effects on colony yields, bacterial species diversity, and cultivation of new species. To reach this objective, culturable cell numbers and species diversities were compared on different agar media. The diversity of the culturable bacterial community from the potato rhizosphere was also compared with that determined by a cultivation-independent technique. A selection of isolates from all agar media was identified by 16S rRNA gene sequencing.
2 Materials and methods
2.1 Rhizosphere sampling and processing
Seven potato plants (Solanum tuberosum L.) cv. Agria, healthy in their appearances, were randomly collected from an agricultural field (250 m2 in size) near Wageningen, The Netherlands (51°59′N, 5°39′E), at flowering stage, growth stage 6 (Hack et al. 1993). Minimal distances between replicate plants was 1 m, and from each individual plant, roots with soil were taken and stored on moisture paper towels in closed boxes for transport to the laboratory where samples were further processed (maximum time between sampling and processing was 1 h).
The soil from the field plot was typed as loamy sand containing 2% organic matter, a water holding capacity of 25%, and a pH (KCl) of 4.8. Soil adhering to the roots of each plant after shaking by hand was considered to represent the rhizosphere. This soil, approximately 1 g, was washed from the roots by shaking for 10 min in 200 ml of 0.1% sodium pyrophosphate solution. The thus obtained rhizosphere soil suspensions were blended for 1 min at 12,000 rpm, and blending was repeated two times—with 30-s intervals—to release as many bacterial cells as possible. Rhizosphere suspensions were used for dilution plating, cell counting, and total DNA extraction followed by polymerase chain reaction (PCR) amplification and construction of the rhizosphere DNA library.
2.2 Agar medium preparation and plating
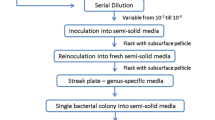
R2A (Difco™, France) was prepared according to the procedure described by the manufacturer. Oligotrophic agar medium was prepared in accordance with Semenov et al. (1999), with the following modifications: purified agar was used instead of Noble agar, and the total carbon content was ten times lower in our agar medium. The following ingredients were dissolved in 1 L of Millipore-membrane filtered ultrapure water: MgSO4, 0.5 g; KNO3, 0.5 g; K2HPO4, 1.3 g; Ca(NO3)2, 0.06 g; glucose, 0.05 g; hydrolyzed casein, 0.004 g; purified agar (Oxoid, Basingstoke, UK), 12 g (pH 7.5); followed by sterilization at 121°C for 20 min. Molten oligotrophic agar was first allowed to cool down to 47°C, after which, it was either mixed with catalase (CAT; 100 U per milliliter, Sigma, The Netherlands) or left unmixed (OLI). Potato rhizosphere extract agar was prepared by mixing molten oligotrophic agar without glucose (47°C) with potato rhizosphere extract reaching a final carbon concentration of 0.05 g per liter (PEX). Potato rhizosphere extract was made from potato rhizosphere soil by incubation of 20 g of potato roots with tightly adhering soil in 1 L of sterilized Millipore-membrane-filtered ultrapure water overnight at 4°C. Then, soil particles were removed by centrifugation for 10 min at 200 g, and the remaining supernatant was sterilized by filtration through a 0.22-μm filter (MILLEX® GP, Millipore, Ireland; Da Rocha et al. 2009). Sterility of thus obtained filtrate was checked by spreading 100 µl onto OLI plates and incubation in moisturized boxes at 25°C for 15 days; this resulted in the absence of any colony formation.
2.3 Bacterial cell enumerations
For total bacterial cell enumeration in the potato rhizosphere, cells in soil suspensions were stained with 5-([dichlorotriazinyl]amino) fluorescein (DTAF; Sigma-Aldrich, St. Louis, MO, USA) followed by automated counting using confocal microscopy, performed according to the procedure of Bloem et al. (1995). For CFU enumeration from the potato rhizosphere, the rhizosphere suspensions were serially tenfold diluted in sterile distilled water, after which (per dilution), 0.1 ml was plated onto R2A, OLI, CAT, or PEX media. Plates kept in moisturized boxes were incubated at 25°C, and colony formation was followed in time by counting the number of CFU between 3 and 15 days. Colonies were distinguished upon visibility by the unarmed eye (colonies, CFU, >250 µm in diameter) or at 50 times magnification (microcolonies, mCFU, between 50 and 250 µm in diameter). mCFUs were enumerated by scanning entire agar plates under aseptic conditions in a laminar flow hood to circumvent contamination from the air. Total DTAF-stainable (fluorescence-stained) cell and CFU numbers were expressed per gram of dry rhizosphere soil and log-transformed prior to statistical comparisons.
2.4 Colony selection, genomic comparison by BOX-PCR fingerprinting, and storage of isolates
After 15 days of incubation, a total of 180 (45 from each agar medium) CFUs were randomly picked from plates that had received the two highest dilutions of the rhizosphere soil suspensions. CFUs were streaked to purity on the same agar medium and allowed to grow out to new colonies after 5 days of incubation. mCFUs were picked from the agar surfaces by making use of sterile dissecting needles under 50 times magnification in a sterile environment. A total of 12 (four from each agar medium: OLI, CAT, and PEX) mCFUs were selected, and cell material from single colonies were transferred to fresh agar media of their respective composition to allow formation of new colonies after 150 days of incubation.
A total of 180 CFUs plus one resulting from an mCFU were compared by BOX-PCR (Rademakers et al. 1998) fingerprinting. Volumes of 1 µl cell lysate, prepared from material from single colonies suspended in 100 µl of sterile demineralized water and heated to 100°C for 5 min, were used as DNA templates for PCR reactions. After PCR amplifications, volumes of 10 μl of each mixture were loaded on 1.2% agarose gels, 40 cm in sizes, and run for 16 h at 4°C and 1.2 V/cm. Each individual gel included three lanes, each loaded with 5 μl of 1 kb ladder (Invitrogen, Carlsbad, CA, USA), for later normalization. After running, gels stained with ethidium bromide were photographed under UV, and digitized fingerprints were compared using Gelcompare II software (Applied Maths, Belgium). BOX-PCR fingerprints were compared at 100%, 95%, and 90% similarity levels.
For storage of isolates, cells from single colonies were suspended in oligotrophic broth (OLI with omission of agar) amended with glycerol (final concentration 20%) and placed at −80°C.
2.5 Construction of a 16S rRNA gene clone library made from rhizosphere DNA
A 16S rRNA gene clone library was made from potato rhizosphere soil. Therefore, DNA was extracted from a pooled suspension from all seven rhizospheres using the UltraClean Soil DNA Isolation Kit (MO BIO laboratories, Inc., Carlsbad, CA, USA). Then, 1 µl (20 ng) of purified DNA extract was PCR amplified using primers 27F and 1492R. The PCR product with the expected fragment size was purified from non-incorporated dNTPs and primers using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Purified PCR product was cloned into the pCR2.1-TOPO vector from the TOPO-TA PCR cloning kit (Invitrogen) and introduced into Escherichia coli Top10 cells (Invitrogen) by transformation according to the protocol provided by the manufacturer. White colonies, indicating insertional inactivation of the lacZ gene, were PCR amplified with primers annealing with the M13F and M13R sites in the pCR2.1-TOPO vector using the procedure provided by the manufacturer. In total, 93 clones with the expected fragment size were selected for later sequencing.
2.6 Identification of isolates and clone inserts from the rhizosphere DNA library by 16S rRNA gene sequencing
For PCR amplification of the 16S rRNA gene, cell lysates from all 180 isolates recovered as CFUs and 12 as mCFUs (192 in total) were used as DNA templates. For that, cell material was used from purified colonies with the exception of 11 mCFUs that were not able to grow upon transfer to fresh media, and from these, cell material taken from the original colonies were added to the PCR mix. Per cell lysate, 1 µL was added to 49 µL PCR reaction mixtures consisting of Tris–HCl (pH 8.3), 10 mM; KCl, 10 mM; MgCl2, 2.5 mM; each deoxyribonucleoside triphosphate, 200 mM; 400 mM of each primer, 27F (Lane et al. 1985) and 1492R (Rochelle et al. 1992), and 5 U of SuperTaq DNA Polymerase (HT Biotechnology, UK). PCR amplifications were run in a PTC-200 thermal cycler (MJ Research, The Netherlands) programmed at one cycle of 95°C, 5 min; 30 cycles at 95°C for 60 s, 62°C for 60 s, and 72°C for 90 s; and one cycle 72°C for 10 min. The PCR products obtained were used for later sequencing.
PCR products of all 192 isolates from the four agar media and the 93 clones from the rhizosphere DNA library were sequenced. For that purpose, purified PCR products were added to reaction mixtures containing 5 μl of sequencing reaction mixture, 1 µl of DETT Dye (Dyenamic ET Terminator Cycle Sequencing Kit, Healthcare, GE), 3 μl of dilution buffer, and 1 µl (0.5 μM) of primer 1492R. Linear amplifications were performed for 25 cycles at 94°C, 20 s; 50°C, 15 s; and 60°C, 60 s. The amplified products, approximating 600 bp in size, were sequenced in an ABI prism automatic sequencer by making use of the services of Greenomics (Plant Research International, Wageningen, The Netherlands). Sequence data from all isolates and clones were first checked for chimeras using the Check Chimera tool (http://www.ncbi.nlm.nih.gov), and nonchimeric sequences were assessed for similarity, using the Sim_Identity index (SI) with sequences of type strains in the Ribosomal Database Project (RDP; Cole et al. 2009) using the bioinformatic analysis tools of the Sapelo Island Microbial Observatory (last accession 30 September 2009; http://simo.marsci.uga.edu/). The identities of colonies and clones, expressed in operational taxonomic units (OTUs), were provided according to their nearest matches at levels of 99% (strain), 95% (genus), and 90% (class) with sequences present in the database. OTUs of 12 isolates showed <95% similarity with sequences of type strains in the RDP, and these isolates were considered as non-readily identifiable. Sequencing of a larger part of the 16S rRNA genes of these isolates was performed for better comparisons with SILVA release 94 (Pruesse et al. 2007) database entries. The following primers were used for further sequencing of these 16S rRNA genes: P027F (Lane et al. 1985), R530 (Muyzer et al. 1993), and 968R (Heuer et al. 1997). Assembled contiguous fragments were compared for nearest matches with SILVA database entries using ARB software (Ludwig et al. 2004).
2.7 Data analysis
For statistical analyses, comparisons were made between total fluorescence-stained cell and total CFU numbers from all four agar media, between total CFU numbers from the different agar media after 15 days of incubation. Differences were calculated by Tukey's test with seven replicates per sample and were considered to be significant at a level of P ≤ 0.05.
Rarefaction analysis was performed on isolates of the four media and clones of the rhizosphere DNA library to determine OTU richness at 99% and 95% similarity levels. The rarefaction modus of the DOTUR software (http://www.plantpath.wisc.edu/fac/joh/dotur.html) was used (Schloss and Handelsman 2005). Rarefaction diagrams were made by plotting the number of OTUs as a function of the number of individual colonies or clones sampled from, respectively, the different media or the rhizosphere DNA clone library.
Correspondence between pools of (V6–V8) rRNA gene sequences from library clones and isolates was made by pairwise comparisons using ∫-LIBSHUFF (Schloss et al. 2004). Therefore, sequences from different pools were aligned, and a matrix of similarity was generated by the application of DNADist, which was used as input for analysis by ∫-LIBSHUFF, after which, corrected P values were calculated.
Phylogenetic distances were calculated using the ARB software package between 16S rRNA gene sequences of over 1,000 bp in size from four selected isolates and 90 other Verrucomicrobia sequences and ten representatives from the phylum of Chlamydiae as an out group from the Silva database. Aligned sequences were manually edited taking 16S rRNA sequence secondary structures into consideration. Reconstruction of phylogenetic relationships was based on neighbour joining (Ludwig et al. 1998). The branches were tested with bootstrap analysis (1,000 iterations).
2.8 Nucleotide sequence accession numbers
The DNA sequences of the partial 16S rRNA genes of 12 isolates (between 804 and 1,492 bp in sizes) were deposited in the EMBL Nucleotide Sequence Database (Cochrane et al. 2009) under accession numbers FN394501 to FN394512.
3 Results
3.1 Bacterial numbers in the potato rhizosphere
The log-transformed fluorescence-stained cell numbers from seven potato rhizospheres, expressed per gram of dry rhizosphere soil, were between 9.19 and 9.58 (Table 1). CFUs from the same samples appeared on the R2A, OLI, CAT, and PEX media within 7 days of incubation (Fig. 1). This stood in contrast to the microcolonies (mCFU), which in general appeared later, i.e., between 7 and 15 days of incubation (see Fig. 1). After 15 days of incubation, the average log CFU per gram of dry soil was 8.09 on R2A, 8.27 on OLI, 8.55 on CAT, and 8.36 on PEX (see Table 1). The log mCFU per gram of dry soil were 8.21 on OLI, 8.39 on CAT, and 8.45 on PEX. No mCFU were found on R2A. Log total CFU (CFU + mCFU) per gram of dry soil after 15 days of incubation were 8.09 on R2A, 8.54 on OLI, 8.78 on CAT, and 8.71 on PEX. These numbers were roughly between 0.47 and 1.58 log units lower than the corresponding total cell counts as determined by fluorescence-stained cell numbers. Expressed as percentages of the fluorescence-stained cell numbers, the total CFUs on day 15 were between 2.6% and 6.6% on R2A, 4.5–33.9% on OLI, 14.4–33.6% on CAT, and 12.3–28.1% on PEX. The sizes of the culturable fractions thus differed per medium and incubation time and occasionally comprised (highest value found on OLI medium) almost one third of the total fluorescence-stained cell fraction. Considering variability over the different plants, the isolate fractions across the different agar media were lowest in plant 7 and highest in plant 6. Hence, the sizes of these fractions in the rhizosphere varied among the replicate plants. The medium used for the recovery of bacterial cells from the potato rhizosphere was determinative for the level of culturability. The highest isolate numbers were found on CAT, PEX, and OLI media, whereas such numbers were significantly lower on R2A (see Table 1).
3.2 Successive growth of isolates on agar media
A total of 192 isolates obtained from both CFUs and mCFUs were successively streaked to purity and transferred to fresh media in order to determine their capacity to show continued growth under medium conditions. Isolation of cells from mCFU is a laborious process and, hence, we included a maximum of 12 such isolates. Thus, 180 isolates (45 from each agar medium) were obtained from CFUs and 12 (four from each agar medium: OLI, CAT, and PEX) from mCFUs. All isolates recovered as CFUs grew into CFUs upon transfer to the same agar medium, whereas 11 of the isolates recovered as mCFUs did not form mCFUs or CFUs, not even after 150 days of incubation. The one mCFU (from CAT, denoted as CR28) that did grow formed a CFU after transfer to the same agar medium within 5 days of incubation. Hence, whereas all CFUs maintained culturability, the majority of mCFU did not.
3.3 Comparison of BOX-PCR fingerprints from culturable isolates
Cell lysates prepared from 181 isolates (i.e., the ones that grew upon transfer) were analyzed by BOX-PCR to cross-compare their genomes (data not shown). Similarity levels of 100%, 95%, and 90% were used in the groupings. At 100% and 95% of similarity, all isolates differed from each other, indicating that a huge diversity of culturable forms was indeed sampled. At 90% similarity, three clusters, consisting of respectively two, five, and six isolates, were distinguished, whereas all other 168 fingerprints formed singletons that differed from each other. No commonalities among isolates from the same agar medium were found at this level. This indicates that all isolates differed substantially from each other, irrespective of their initial growth on different agar media.
3.4 Comparison of partial 16S rRNA gene sequences of isolates with database entries
The sequences of the variable regions V6–V8 of the 16S rRNA genes of 180 isolates from all four agar media were affiliated with, in total, five different phyla. Representatives of the Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes were obtained from all four media, but those of the Verrucomicrobia only from CAT and PEX (Table 2). Differences in the dominant groups on the four media were found at the class level. On R2A, the group of α-Proteobacteria was the largest, on OLI the Flavobacteria and α-Proteobacteria, on CAT the Flavobacteria, and on PEX the α-Proteobacteria and Actinobacteria. The α-Proteobacteria was the most dominant class among isolates from R2A and PEX, whereas Flavobacteria were dominant among the ones from OLI and CAT.
In total, 169 (of 180) sequences of isolates recovered as CFUs from all four agar media matched database entries of type strains at 95% or higher levels, as follows: 44 from R2A, 41 from OLI, 41 from CAT, and 42 from PEX. Eleven isolates showed similarities at levels <95% with type strains of the RDP. These isolates were regarded as non-readily identifiable, and so, larger stretches of their 16S rRNA genes were sequenced for more accurate comparisons with database entries (Table 3).
Partial 16S rRNA gene sequences of the 12 isolates recovered as mCFUs were affiliated with four phyla, i.e., the Bacteroidetes (class Flavobacteria), Actinobacteria (class Actinobacteria), Proteobacteria (class α-Proteobacteria), and Verrucomicrobia (subdivision 1; see Table 2). The 16S rRNA gene sequence of the isolate that showed stable growth when streaked on the same medium, CR28, closely matched those of the subdivision 1 of Verrucomicrobia. The aforementioned four taxonomic groups were not all recovered from different agar media, indicating that unique selection of particular bacterial groups by the different agar media had not occurred.
Only one isolate recovered as an mCFU closely matched the 16S rRNA gene sequence of a type strain (Flavobacterium hydatis) in the RDP database at a similarity level >99% (Table 4). At similarity levels between 99% and 95%, the 16S rRNA genes of in total four isolates were affiliated with those of type strains and eight with those of non-type strains. At similarity levels <95%, another four isolates (amongst which CR28) were affiliated with non-type strains, indicating that these were only distantly related to representative species present in the RDP.
3.5 Potato rhizosphere DNA library analysis and comparison between cultured and uncultured bacteria
A total of 93 PCR-amplified 16S rRNA gene sequences from uncultured bacteria was obtained from the potato rhizosphere. DNA sequence analysis of all clones revealed that there were no distinguishable groups at a 97% similarity level (meaning all sequences were singletons), whereas at a 95% similarity level, only two clusters, both consisting of two sequences, were distinguished. Hence, most of the clones in the rhizosphere 16S rRNA gene library were indeed singletons, indicating a high diversity within the bacterial community that was sampled.
In total, only ten of the 93 sequences matched type strains in the RDP database at similarity levels >95%, as follows: Streptomyces ciscaucasicus (99.2%), Arthrobacter ramosus (99.1%), Variovorax paradoxus (two isolates: 98.6% and 98.2%), Aquabacterium parvum (97.6%), Aquabacterium commune (97.1%), Agrococcus jenensis (96.5%), Terrimonas ferruginea (95.6%), Paenibacillus alkaliterrae (95.2%), and Sphingosinicella microcystinivoran (95.0%). The other 83 sequences showed similarity values <95% to RDP database entries, revealing best matches with the Acidobacteria, Proteobacteria, Actinobacteria, Bacteroidetes, Gemmatimonadetes, Firmicutes, Verrucomicrobia, Planktomycetes, Chloroflexi, division OP10 and unclassified bacteria (see Table 2). Of the two groups that were most abundant in the clone library, i.e., the Acidobacteria and Proteobacteria, nearest matches with particular groups or classes were found: for the Acidobacteria groups 3, 5, 6, and 7 and for the Proteobacteria, α-Proteobacteria, β-Proteobacteria, δ-Proteobacteria, and γ-Proteobacteria.
A comparison between the clone library sequences and those from the 192 isolates by ∫-LIBSHUFF analyses revealed that the two libraries differed more than 5% from each other (P < 0.0001), indicating that the correlation between the two was low. Thus, the sequences present in the directly obtained pool from the potato rhizosphere differed substantially from the ones present in the isolates.
Rarefaction analysis performed at similarity levels of 99% and 95% on the basis of the clone library sequences showed an almost linear relationship between clone number and type (Fig. 2). Hence, an increased sampling from the rhizosphere DNA library would have yielded additional novelty at both similarity levels, and the rhizosphere DNA was not fully explored for diversity. Similarly, rarefaction curves (99% similarity level) based on the 16S rRNA gene sequences of isolates obtained from the OLI and PEX media showed almost linear relationships, whereas those from R2A and CAT tended to level off. This observation indicated that progressively lower numbers of new species are expected to be recovered from the latter media upon further sampling. At 95% similarity levels, the rarefaction curves of 16S rRNA gene sequences of isolates from all four agar media tended to level off.
The sequences obtained in the clone library were aligned with those of the 192 isolates to examine the putative matches between the two sequence pools. At six occasions, matches between the sequences of clones and isolates were found (similarity levels between 95% and 98%). Three library clones showed matches with isolates from OLI medium. Specifically, these clones matched the 16S rRNA gene sequences of A. ramosus, V. paradoxus, and Ramlibacter henchirensis. Two clones showed matches with isolates from R2A, affiliating with Stenotrophomonas acidaminiphila and Sphingobium yanoikuyae, whereas one matched an isolate from PEX medium with closest affiliation to A. jenensis. No sequences that were identical between the clone library and any of the isolates from the four media were found. This emphasizes the difference between the cultured and uncultured fractions obtained from the potato rhizosphere.
3.6 Identity of culturable bacteria with less than 95% similarity to RDP database entries
The sequences of, in total, 12 isolates (11 recovered as CFUs and one as a mCFU) showed <95% similarity with sequences of type strains of the RDP database. These isolates should hence be considered as novel taxa. The isolates had been obtained from R2A (1), OLI (4), CAT (4), and PEX (3). Larger (804–1,114 bp) 16S rRNA gene sequences of these isolates were obtained and analyzed using sequences from the RDP, thus allowing more accurate identifications. However, rather distant nearest matches were found with (type strains) Rubritalea marina (four isolates; nearest matches between 88.4% and 89.6%), Pedobacter roseus (three; 85.1% and 94.2%), Arthrobacter nitroguajacolicus (one; 93.1%), Lysobacter antibioticus (one; 92.1%), Pantoea agglomerans (one; 90.0%), Sphingobium xenophagum (one; 93.3%), and Methylobacterium rhodinum (one; 93.2%; see Table 3). All matches to type strains were clearly below 95%. However, some sequences matched those of non-type strains from the Silva database. Remarkable were the matches of the four isolates CR28, C20, Z35, and ZNBB5 (which matched that of the R. marina type strain at 88.9% or below) with unidentified/uncultured bacteria at 96.5% similarity or higher. Furthermore, the four isolates clustered with four uncultured representatives of subdivision 1 of the Verrucomicrobia, as shown in a dendrogram constructed with sequences of representatives of the seven subdivisions of Verrucomicrobia (Fig. 3). Given the fact that the isolates originated from CAT and PEX, these two media may allow the enhanced isolation of representatives of subdivision 1 of Verrucomicrobia.
Phylogenetic relationship between four non-readily identifiable isolates, R28, C20, Z35, and ZNBB5 and representatives from the phylum Verrucomicrobia. Distances between partial 16S rRNA gene sequences over 1,100 bp in length were calculated using ARB software. A cluster made of rRNA gene sequences of ten different species from the phylum Chlamydiae was used as an out group. Bar at the bottom indicate 10% divergence among sequences, and boot strap values (percent) are presented near each junction in the tree
On the other hand, no 16S rRNA gene sequences were found in the clone library that matched representatives of subdivision 1 of Verrucomicrobia (Silva database). However, two clone library sequences showed nearest matches with representatives of subdivision 3 of the Verrucomicrobia. One matched an uncultured soil bacterium affiliated with unclassified Verrucomicrobia (accession number AY493907) and the other one an uncultured Verrucomicrobium sp. (AY921923). The similarity levels were, respectively, 98.7% and 99.0%. Representatives of subdivision 3 of the Verrucomicrobia may be more dominant in the potato rhizosphere than those of subdivision 1, whereas only the latter could be recovered as isolates on CAT and PEX.
4 Discussion
4.1 Improvement of culturability
A comparative study on the recovery of bacteria from the potato rhizosphere on different agar media was conducted. The total CFU numbers that were found occasionally contributed to substantial fractions (up to 33.6%) of the total fluorescence-stained cell counts, depending on the type of medium used, incubation time, and the presence of mCFU and plant replicate. Our data thus show that culturability from the rhizosphere can be substantially improved by using oligotrophic agar media with or without modifications in comparison with R2A, representing a standard laboratory medium.
The agar media in this study were employed with the rationale to enhance the bacterial culturability by reducing cellular stresses upon exposure to oxygen (catalase) or high-nutrient conditions, the latter by mimicking the (low) nutritional status of the rhizosphere (rhizosphere extract). The presence of catalase reduces oxidative stress at the agar surface resulting in better growth and higher CFU yields (Barer and Harwood 1999; Bogosian and Bourneuf 2001; Wagner and Horn 2006). We indeed found higher CFU numbers on catalase-containing agar medium, however, at the expense of a lowered diversity, both compared to those on the corresponding unamended agar. The lowered diversity may have been caused by a faster outgrowth of colonies on CAT, which was apparent from the larger colony sizes, than on OLI medium (not shown). Faster-growing colonies may suppress the growth of slower ones on the same plate (Kamagata and Tamaki 2005), which is consistent with our observations.
Mimicking the nutritional status of the rhizosphere was indeed successful concerning the recovery of novel bacterial types, as previously also found for bulk soil (Hamaki et al. 2005). Rhizosphere soil is distinguished from bulk soil as an environment differing in species composition (Van Overbeek and van Elsas 2008), which incites a higher microbial activity as a result of the availability of plant root exudates (Van Overbeek and van Elsas 2008; Wagner and Horn 2006). By replacing glucose as the main carbon source by rhizosphere extract (which in reality is a combination of root exudate and soil-extractable compounds), the growth of rhizosphere bacteria was expected to be favored. Higher total CFU numbers were indeed found on PEX than on OLI medium. Moreover, the slight difference in composition of bacterial groups between the two media reflected the variation in the preference for carbon sources among bacteria present in the potato rhizosphere.
The presence of the mCFUs on the media made up large part of the observed higher CFU numbers. Concerning the physiological status of the bacterial cells that grew in these mCFUs, it becomes apparent that the majority only divided for just the number of generations it took to form the mCFU. Possibly, these cells suffered from substrate-accelerated death in the trajectory from rhizosphere soil via one medium to the next medium. In previous work, microcolonies have been obtained from water and soil (Kaeberlein et al. 2002; Zengler et al. 2002; Ferrari et al. 2005), and the bacteria forming these colonies represented, for the greatest part, as-yet-uncultured groups. For instance, a representative of candidate division TM7 was found in soil (Ferrari et al. 2005). mCFUs may represent an untapped resource for many novel species, and this stresses their importance as sources of unexplored bacteria from soils. However, the lack of stable growth upon transfer awaits a future solution.
The large variation in the recovered culturable fractions per plant indicates that the circumstances for efficient growth of different bacterial groups vary per plant, even under almost identical conditions regarding soil type, plant genotype, and growth conditions. Stochasticity in bacterial colonization of the rhizosphere is one factor that may explain this large variation in uncultured fractions (66–96%) in the rhizosphere. Cells of these uncultured bacteria may have been either viable but unable to grow outside the rhizosphere, or moribund and thereby unable to divide.
4.2 Attempts to cultivate hitherto-uncultured representatives of Acidobacteria and Verrucomicrobia from the rhizosphere
Improvements of culturability have been described before for soil (Janssen et al. 2002; Schoenborn et al. 2004) and also freshwater (Bruns et al. 2003). These have resulted in the isolation of bacteria that were previously known for their recalcitrance for growth, like those belonging to the Acidobacteria (Janssen et al. 2002). In our study, the approaches used did not result in the cultivation of members of the Acidobacteria, but our sampling was obviously limited. On the other hand, the recovery of six different acidobacterial groups in the clone library indicates that representatives of this phylum are quite abundant in the potato rhizosphere. Acidobacteria, especially members of group 6, have been found in rhizospheres of other plant species (Schmalenberger and Tebbe 2003; Sharma et al. 2005; Wang et al. 2007; Hao et al. 2008; Kielak et al. 2009). Acidobacteria, albeit quite diverse, as a group has been shown to thrive in bulk soils and also in the rhizosphere (Buée et al. 2009; Nunes da Rocha et al. 2009).
Representatives of another bacterial phylum recalcitrant for cultivation, the Verrucomicrobia, were also found. The organisms found among the isolates differed from those found in the total fraction, i.e., at the DNA level. The isolates belonged to subdivision 1, whereas the DNA-based evidence pointed to subdivision 3 Verrucomicrobia. The isolates matched R. marina but showed substantially higher affiliations with either an unidentified or an uncultured bacterium. R. marina is often found in sponges (Scheuermayer et al. 2006), the unidentified bacterium in the Agrostis stolonifera (creeping bentgrass) rhizosphere (http://turf.lib.msu.edu/tero/v03/n24.pdf; September 28, 2009), and an uncultured bacterium in sediment (Nelson et al. 2007). They thus resemble hitherto-undescribed verrucomicrobial species, one of which also originated from rhizosphere soil. The four isolates grouped together in a cluster of species belonging to the subdivision 1 Verrucomicrobia, indicating that they belong to a separate branch within this phylum.
It was not expected on beforehand that subdivision 1 Verrucomicrobia were present among the isolates in such high frequency. To the best of our knowledge, subdivision 1 Verrucomicrobia isolates have never been obtained from the rhizosphere. The abundance of subdivision 1 Verrucomicrobia among potato rhizosphere isolates indicates that they may thrive in this habitat, although their relationship with the plant roots is so far unexplored. Their isolation on CAT and PEX media indicates that both media offer conditions propitious for growth.
So far, the Verrucomicrobia encompass cultured as well as uncultured species (Hedlund et al. 1997; Stevenson et al. 2004; Sanguin et al. 2006; Wagner and Horn 2006). Most observations of verrucomicrobial species in soils are from direct molecular studies. Verrucomicrobial species often show relative dominance in soils, as up to 9.8% of the total extractable RNA from, e.g., arable soil can consist of verrucomicrobial rRNA (Buckley and Schmidt 2001; Wagner and Horn 2006). It is unclear whether Verrucomicrobia prefer bulk or rhizosphere soils, and reports on their prevalence are occasionally contradictory (Buckley and Schmidt 2001; Sanguin et al. 2006; Kielak et al. 2008). Although we ignore to which extent members belonging to Verrucomicrobia dominate the bacterial community in the potato rhizosphere, it can be concluded that culturable representatives belonging to subdivision 1 and the uncultured ones belonging to subdivision 3 of the Verrucomicrobia are present in the potato rhizosphere. Contradicting information about the prevalence of Verrucomicrobia in this habitat may be based on general statements about the entire phylum and not on the individual subdivisions within this phylum.
5 Conclusions
In conclusion, cultivation was improved, and isolate fractions as high as one third of the total microscopically discernable bacterial cells from the potato rhizosphere were obtained. Application of oligotrophic media (either or not amended with potato root exudates or catalase) in combination with selection for mCFUs resulted in raised CFU numbers. However, improved cultivation did not always yield a higher taxonomical diversity of culturable bacteria. Comparison between the isolate and directly detectable bacterial fractions from the potato rhizosphere revealed that there was still a large difference between both.
A major finding in this study was the isolation of subdivision 1 Verrucomicrobia from the potato rhizosphere. Isolates of this group have not yet been recovered from rhizosphere soil. Considering the relatively high number of isolates belonging to this group, we conclude that the Verrucomicrobia subdivision 1 is an important member of the bacterial community in the potato rhizosphere. Its occurrence in the highest-dilution soil suspensions indicates that it numerically dominate the communities of the potato rhizosphere.
6 Recommendations and perspectives
New isolates of Verrucomicrobia subdivision 1 will be further investigated for their ecophysiological roles in the rhizosphere. Important questions that will be answered in follow-up studies are (1) to which extent do members of this group prevail in the rhizosphere and (2) what is their relationship with the plant roots and their associated communities. These studies will provide a new insight in the roles of newly cultured bacteria that are thought to dominate plant/soil ecosystems. Thus, new mechanistic effects on plant growth and development may be found, which is difficult to achieve with culture-independent approaches alone.
References
Barer MR, Harwood CR (1999) Bacterial viability and culturability. In: Poole R (ed) Advances in microbial physiology 41. Academic, New York, pp 93–137
Bloem J, Veninga M, Shepherd J (1995) Fully-automatic determination of soil bacterium numbers, cell volumes, and frequencies of dividing cells by confocal laser-scanning microscopy and image-analysis. Appl Environ Microbiol 61:926–936
Bogosian G, Bourneuf EV (2001) A matter of bacterial life and death. EMBO Rep 2:770–774
Bruns A, Nubel U, Cypionka H, Overmann J (2003) Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl Environ Microbiol 69:1980–1989
Buckley DH, Schmidt TM (2001) Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol Ecol 35:105–112
Buée M, De Boer W, Martin F, van Overbeek L, Jurkevitch E (2009) The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil 321:189–212
Cochrane G, Akhtar R, Bonfield J et al (2009) Petabyte-scale innovations at the European nucleotide archive. Nucleic Acids Res 37:D19–D25
Cole JR, Wang Q, Cardenas E et al (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Curl AE, Truelove B (1986) The rhizosphere. Springer-Verlag, Berlin
Da Rocha UN, Tótola MR, Pessoa DMM, Júnior JTA, Neves JCL, Borges AC (2009) Mobilisation of bacteria in a fine-grained residual soil by electrophoresis. J Hazard Mater 161:485–491
Eilers H, Pernthaler J, Glockner FO, Amann R (2000) Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol 66:3044–3051
Felske A, Wolterink A, van Lis R, de Vos WM, Akkermans ADL (1999) Searching for predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol Ecol 30:137–145
Ferrari BC, Binnerup SJ, Gillings M (2005) Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol 71:8714–8720
Guan LL, Kamino K (2001) Bacterial response to siderophore and quorum-sensing chemical signals in the seawater microbial community. BMC Microbiol 1:1–11
Hack H, Gall H, Klemke T, Klose R, Meier U, Stauss R, Witzenberger A (1993) Phänologische Entwicklungsstadien der Kartoffel (Solanum tuberosum L.). Nachrichtenbl Deut Pflanzenschutzd 45:11–19
Hamaki T, Suzuki M, Fudou R, Jojima Y, Kajiura T, Tabuchi A, Sen K, Shibai H (2005) Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J Biosci Bioeng 99:485–492
Hao DC, Ge GB, Yang L (2008) Bacterial diversity of Taxus rhizosphere: culture-independent and culture-dependent approaches. FEMS Microbiol Lett 284:204–212
Hedlund BP, Gosink JJ, Staley JT (1997) Verrucomicrobia div. nov., a new division of the bacteria containing three new species of Prosthecobacter. Antonie van Leeuwenhoek 72:29–38
Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241
Ingham CJ, Sprenkels A, Bomer J, Molenaar D, van den Berg A, van Hylckama Vlieg JET, de Vos WM (2007) The micro-petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc Natl Acad Sci USA 104:18217–18222
Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M (2002) Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl Environ Microbiol 68:2391–2396
Kaeberlein T, Lewis K, Epstein SS (2002) Isolating ‘uncultivable’ microorganisms in pure culture in a simulated natural environment. Science 296:1127–1129
Kamagata Y, Tamaki H (2005) Cultivation of uncultured fastidious microbes. Microbes Environ 20:85–91
Kielak A, Pijl AS, van Veen JA, Kowalchuk GA (2008) Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol Ecol 63:372–382
Kielak A, Pijl AS, van Veen JA, Kowalchuk GA (2009) Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J 3:378–382
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR (1985) Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA 82:6955–6959
Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer KH (1998) Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554–568
Ludwig W, Strunk O, Westram R et al (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl Environ Microbiol 59:695–700
Nelson DM, Ohene-Adjei S, Hu FS, Cann IKO, Mackie RI (2007) Bacterial diversity and distribution in the Holocene sediments of a northern temperate lake. Microb Ecol 54:252–263
Nunes da Rocha U, van Overbeek L, van Elsas JD (2009) Exploration of hitherto uncultured bacteria from the rhizosphere. FEMS Microbiol Ecol 69:313–328
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rademakers JLW, Louws FJ, de Bruijn FJ (1998) Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting. In: Akkermans ADL, van Elsas JD, de Bruijn FJ (eds) Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, pp 1–27 chapter 3.4.2
Rasche F, Hodl V, Poll C, Kandeler E, Gerzabek MH, van Elsas JD, Sessitsch A (2006) Rhizosphere bacteria affected by transgenic potatoes with antibacterial activities compared with the effects of soil, wild-type potatoes, vegetation stage and pathogen exposure. FEMS Microbiol Ecol 56:219–235
Rochelle PA, Fry JC, Parkes RJ, Weightman AJ (1992) DNA extraction for 16S ribosomal RNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol Lett 100:59–65
Sanguin H, Remenant B, Dechesne A, Thioulouse J, Vogel TM, Nesme X, Moënne-Loccoz Y, Grundmann GL (2006) Potential of a 16S rRNA-based taxonomic microarray for analyzing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl Environ Microbiol 72:4302–4312
Scheuermayer M, Gulder TAM, Bringmann G, Hentschel U (2006) Rubritalea marina gen. nov., sp. nov., a marine representative of the phylum ‘Verrucomicrobia’, isolated from a sponge (Porifera). Int J Syst Evol Microbiol 56:2723–2723
Schmalenberger A, Tebbe CC (2003) Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol Ecol 12:251–261
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schloss PD, Larget BR, Handelsman J (2004) Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol 70:5485–5492
Schoenborn L, Yates PS, Grinton BE, Hugenholtz P, Janssen PH (2004) Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl Environ Microbiol 70:4363–4366
Semenov AM, van Bruggen AHC, Zelenev VV (1999) Moving waves of bacterial populations and total organic carbon along roots of wheat. Microb Ecol 37:116–128
Sharma S, Aneja MK, Mayer J, Munch JC, Schloter M (2005) Characterization of bacterial community structure in rhizosphere soil of grain legumes. Microb Ecol 49:407–415
Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA (2004) New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol 70:4748–4755
Ueda K, Tagami Y, Kamihara Y, Shiratori H, Takano H, Beppu T (2008) Isolation of bacteria whose growth is dependent on high levels of CO2 and implications of their potential diversity. Appl Environ Microbiol 74:4535–4538
Van Overbeek LS, van Elsas JD (2008) Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.). FEMS Microbiol Ecol 64:283–296
Van Overbeek LS, Bergervoet JHH, Jacobs F, van Elsas JD (2004) The low-temperature-induced viable-but-nonculturable state affects the virulence in Ralstonia solanacearum biovar 2. Phytopathology 94:463–469
Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol 17:241–249
Wang M, Chen JK, Li B (2007) Characterization of bacterial community structure and diversity in rhizosphere soils of three plants in rapidly changing salt marshes using 16S rDNA. Pedosphere 17:545–556
Zengler K, Toledo G, Rappé M, Elkins J, Mathur EJ, Short JM, Keller M (2002) Cultivating the uncultured. Proc Natl Acad Sci USA 99:15681–15686
Acknowledgements
This work was supported by the Netherlands Genomics Initiative (Ecogenomics program) and the Dutch Ministry of Agriculture, Nature, and Food quality (KB4, research program on sustainable agriculture). We thank CAPES for the fellowship provided to FDA (process 2990/05-9). We also thank An Vos, Meint Veninga, and Jaap Bloem for their help with total bacterial counts.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Ji-Zheng He
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
da Rocha, U.N., Andreote, F.D., de Azevedo, J.L. et al. Cultivation of hitherto-uncultured bacteria belonging to the Verrucomicrobia subdivision 1 from the potato (Solanum tuberosum L.) rhizosphere. J Soils Sediments 10, 326–339 (2010). https://doi.org/10.1007/s11368-009-0160-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-009-0160-3