Abstract
International conservation goals have been set to mitigate Southern Ocean ecosystem deterioration, with multiple monitoring programs evaluating progress towards those goals. The scale of continuous monitoring through visual observations, however, is challenged by the remoteness of the area and logistical constraints. Given the ecological and economic importance of the Southern Ocean, it is imperative that additional biological monitoring approaches are explored. Recently, marine sponges, which are frequently caught and discarded in Southern Ocean fisheries, have been shown to naturally accumulate environmental DNA (eDNA). Here, we compare fish eDNA signals from marine sponge bycatch specimens to fish catch records for nine locations on the continental shelf (523.5–709 m) and 17 from the continental slope (887.5–1611.5 m) within the Ross Sea, Antarctica. We recorded a total of 20 fishes, with 12 fishes reported as catch, 18 observed by eDNA, and ten detected by both methods. While sampling location was the largest contributor to the variation observed in the dataset, eDNA obtained significantly higher species richness and displayed a significantly different species composition compared to fish catch records. Overall, eDNA read count correlated more strongly with fish abundance over biomass. Species composition correlated on a regional scale between methods, however eDNA signal strength was a low predictor of catch numbers at the species level. Our results highlight the potential of sponge eDNA monitoring in the Southern Ocean by detecting a larger fraction of the fish community compared to catch recordings, thereby increasing our knowledge of this understudied ecosystem and, ultimately, aiding conservation efforts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Marine ecosystems are currently undergoing dramatic shifts in structure and functioning, due to increased anthropogenic impacts and rapid climate change (Allison et al. 2009). Antarctic ecosystems represent some of the least modified marine ecosystems on the planet (Aronson et al. 2011) and the largest marine protected area (Ballard et al. 2012). The Southern Ocean is, however, already being impacted by climate change, notably along the Antarctic Peninsula (Clarke et al. 2006), as well as being exposed to increased fishing and tourism pressures (Aronson et al. 2011; Tejedo et al. 2022). Antarctic-wide impacts of warming, loss of sea ice, and ocean acidification are predicted through modelling over the coming decades (Koerich et al. 2022). While multiple national research programs are currently being undertaken to determine the effects of such a raft of pressures on Antarctica’s marine ecosystem (CCAMLR 2022a, b), Southern Ocean research is by its nature logistically and financially demanding (Xavier et al. 2016). Hence, most regions are understudied and necessary biological information for successful conservation is incomplete (Griffiths 2010; Xavier et al. 2016).
One such data-deprived region is the Ross Sea, a large embayment of the Southern Ocean in Antarctica, between Victoria Land and Marie Byrd Land (74.5487° S, 166.3074° W). The Ross Sea is the southernmost sea on Earth with a total area of 637,000 km2 and exhibits substantial variations in physical forcing, ice cover, and biological processes on a variety of temporal and spatial scales (Smith et al. 2014). The Ross Sea contains some of the most productive waters globally, sustaining the largest phytoplankton biomass in the Southern Ocean (Smith et al. 2014), highly abundant zooplankton (Sala et al. 2002), the most diverse benthos in the Southern Ocean (Clarke and Johnston 2003), and exceptional abundances of apex predators (Ballard et al. 2012). The fish fauna exhibits low diversity and is dominated by notothenioids (cod icefishes), liparids (snailfishes), and zoarcids (eelpouts; Ainley and Pauly 2014; Eastman 2005). The abundance of notothenioids, specifically the Antarctic toothfish (Dissostichus mawsoni Norman, 1937, Nototheniidae), has supported a commercial longline fishery since 1997 (Fisheries New Zealand 2022). While important aspects of the Ross Sea ecosystem are yet to be explored (Griffiths 2010), an international consensus was reached to establish the world’s largest MPA in 2016 to provide protection to previously fished areas (Ballard et al. 2012).
Due to the logistical constraints for routine monitoring in the Southern Ocean, much of the data used to increase our understanding on continental shelf and slope fish population distributions and abundances is generated in association with commercial fishing activities, including catch and bycatch information (Polanowski et al. 2018). These data are integrated into management of fishing activity through the CCAMLR system (Trathan and Agnew 2010). In addition to direct observations of fish catches, the catches provide other opportunities to obtain important biological information associated with the fishery and wider pelagic ecosystems. For example, analysis of gut samples from commercially caught Antarctic toothfish provided new information on their cephalopod prey (Stevens et al. 2014). While essential to our current understanding of the Ross Sea ecosystem, hook-and-line fishing is known to be selective and biased towards the target organism (Løkkeborg and Bjordal 1992; Moreno 1991). Hence, it is imperative that additional monitoring approaches are explored to enable the gathering of essential ecological data.
Environmental DNA (eDNA) monitoring has been proposed as an innovative method with great potential (Ficetola et al. 2008; Thomsen and Willerslev 2015), whereby species are detected indirectly through DNA signals obtained from environmental samples, such as water (Bowers et al. 2021), sediment (Koziol et al. 2019), or air (Lynggaard et al. 2022). Within the marine biome, water is the most frequently used substrate in eDNA surveys (Bowers et al. 2021). Aquatic eDNA surveys have shown to be highly accurate, due to high spatial (Jeunen et al. 2019a, b) and temporal (Berry et al. 2019) resolutions, as well as sensitive, by facilitating early detection of invasive species (Bowers et al. 2021). Furthermore, aquatic eDNA metabarcoding surveys have compared favorably to a variety of traditional monitoring methods with regards to diversity detection in a time-efficient and cost-effective manner (Fediajevaite et al. 2021), including baited remote underwater videos (Jeunen et al. 2020; Stat et al. 2019), trawling (Salter et al. 2019; Stoeckle et al. 2021; Thomsen et al. 2016), and underwater visual census (Polanco Fernández et al. 2021).
Several studies have compared aquatic eDNA metabarcoding to trawling catch records (Salter et al. 2019; Stoeckle et al. 2021; Thomsen et al. 2016). In general, aquatic eDNA tends to recover a larger portion of the fish diversity. While false-negative detections are inherent to all survey methods deployed thus far, false-negative eDNA detections compared to catch records are a frequent occurrence, due to missing reference barcodes (Weigand et al. 2019), low taxonomic resolution in the amplicon region for specific taxonomic groups (Zhang et al. 2020), or amplification bias induced through mismatches in primer-binding regions (Hansen et al. 1998). The partial overlap in species detection, as well as difficulties in obtaining abundance, sex, and size information from eDNA surveys, has led to a proposed combined approach to gather as much information possible (Zhou et al. 2022). Routine implementation of eDNA analysis into existing monitoring programs has so far been hampered by the need for immediate, careful, and time-consuming sample collection, DNA preservation and storage (Bessey et al. 2021).
To circumvent the need for water filtration, passive eDNA collection has been trialed with success (Bessey et al. 2021; Jeunen et al. 2022b; Maiello et al. 2022), whereby filter membranes (Bessey et al. 2021; Jeunen et al. 2022b), artificial sponges (Jeunen et al. 2022b), or other substrates (Maiello et al. 2022; Verdier et al. 2021) are submerged to capture eDNA from the water column. Besides achieving comparable results to active filtration (Jeunen et al. 2022b), passive filtration devices allow for increased sampling and replication by omitting the time-consuming active filtration step (Bessey et al. 2021). An alternative to using artificial substrates for passive eDNA collection is to exploit the natural eDNA accumulation in filter-feeding organisms, such as marine sponges (Cai et al. 2022; Harper et al. 2023; Jeunen et al. 2021; Mariani et al. 2019; Turon et al. 2020). Similar to artificial substrates, sponge eDNA displays high similarity with aquatic eDNA surveys (Jeunen et al. 2021) and might be preferred over using artificial substrates, as they are frequently caught as bycatch and omit the need to attach passive samplers to fishing gear.
In this study, we explore the use of novel eDNA methods to describe the biogeographical patterns of fish on the continental shelf (9 locations; depth range: 523.5–709 m) and slope (17 locations; depth range: 887.5–1611.5 m) in the Ross Sea, Antarctica. Environmental DNA was obtained from marine sponges caught as bycatch on the demersal longline fishing vessel, FV San Aotea II. Catch records enabled us to validate eDNA signals, as well as determine the utility of eDNA biodiversity monitoring in the Southern Ocean. Furthermore, catch biomass and abundance measures were correlated to eDNA signal strength to investigate the quantitative value of eDNA metabarcoding on a regional and local scale. Two questions were specifically addressed: (1) Does the eDNA accumulated in sponges caught as bycatch in longline fisheries detect all caught species and provide additional information to fish biodiversity patterns in the Ross Sea, and (2) can we estimate catch and bycatch biomass/abundance from eDNA obtained from marine sponge bycatch specimens?
Materials and methods
Study area and sample collection
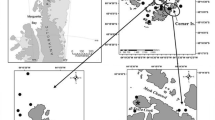
Data between catch records and eDNA detections were compared across a total of 26 sites located on the continental slope (17 locations) and shelf in the Ross Sea (9 locations; Fig. 1; Supplemental Table 1). The exact location of sampling sites has not been disclosed to preserve commercial interest of the fishing vessel.
a Map of Antarctica and the Southern Ocean with sample collection sites in the Ross Sea indicated by a black square. Bathymetry of the Southern Ocean floor is color coded from light blue (shallow) to dark blue (deep sea). Bathymetry information was gathered from Quantarctica inside QGIS (https://www.scar.org/resources/quantarctica/). b Map of Ross Sea and the Ross Sea ice shelf (grey) with the continental shelf sampling region indicated in red and the continental slope sampling region indicated in blue. Fish silhouettes represent the results of the indicator species analysis, with continental shelf eDNA and catch indicator species depicted in light red and dark red, respectively. Continental slope eDNA and catch indicator species are depicted in light blue and dark blue, respectively. Number inside fish silhouettes indicates species name as found in Supplemental Table 8
Sponge specimens were collected during longline fishing by FV San Aotea II on the continental shelf and slope regions of the Ross Sea between the 13th of December 2021 and 17th of January 2022 (Fig. 1; Supplemental Table 1). During the deployment of the longlines, sponges are accidentally hooked off the seafloor and brought to the surface as reported bycatch when the lines are retrieved. A total of 30 marine sponge specimens on 26 fishing lines were sampled for this experiment, with 23 fishing lines represented by a single marine sponge specimen, two fishing lines represented by two marine sponge specimens, and one fishing line represented by three marine sponge specimens. Marine sponges were taxonomically identified to class level (22 Demospongiae [Demosponges]; 8 Hexactinellida [Glass sponges]) on the fishing vessel by fishery observers and each placed in a separate 50 ml falcon tube filled with 99.8% molecular-grade ethanol (Fisher BioReagents™, Fisher Scientific). Specimens were stored in the dark on ice during shipment to the University of Otago’s PCR-free eDNA facilities at Portobello Marine Laboratory (PML). The ethanol-stored specimens were stored at 4 °C in the dark until further sample processing. Due to logistical difficulties of working onboard a commercial fishing vessel in the Southern Ocean, no negative field controls were collected.
Fish catch recordings
For each of the 26 fishing lines where sponges were caught as bycatch, catch composition was recorded by fisheries observers, as per governmental regulations. Although taxon identification is usually conducted at a level coarser than the species—except for toothfish—observers were asked by CCAMLR to identify catches to the lowest taxonomic level possible and measure up to ten individual bycatch species per longline set. Fish bycatch measurements followed standard practices according to the Ross Sea data collection plan (Hanchet et al. 2015) and CCAMLR observer protocols (CCAMLR 2023), which consisted of length, weight, sex, and maturity stage for each recorded specimen. A tissue sample from each species caught on the 26 fishing lines within the taxonomic group Actinopterygii was dissected and shipped frozen to the University of Otago for barcoding purposes. Tissue samples from Chondrichthyes, which are tagged and released after capture, were not obtained for this study.
Fish reference barcodes
A tissue biopsy of ~ 25 mg was dissected from each Actinopterygii species caught as bycatch and extracted using the Qiagen DNeasy Blood & Tissue Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s recommendation, except for an overnight lysis step. Total DNA was quantified using qubit (Qubit™ dsDNA HS Assay Kit, ThermoFisher Scientific) and visualized with gel electrophoresis to determine high molecular weight DNA was present. DNA was amplified using the 16SarL/16SbrH primer set (Palumbi 1991; Supplemental Table 2) to generate reference barcodes for the fish metabarcoding assay used in this experiment. PCR was carried out in 20 µl reactions using BIOTAQ™ DNA Polymerase (Meridian Bioscience®) according to the manufacturer’s instructions, with a 0.2 mM final concentration of each primer and dNTPs, as well as a 2 mM final concentration of MgCl2. The thermal cycling profile included an initial denaturation step of 94 °C for 2 min; followed by 30 cycles of 1 min at 94 °C, 90 s at 53 °C, 90 s at 72 °C; and a final extension step for 10 min at 72 °C. PCR products were checked for amplification through gel electrophoresis, and upon successful amplification the reaction was cleaned using a QIAquick PCR purification Kit (Qiagen, Cat. No. 28104). Cleaned up products were then quantified by spectrophotometry using a DeNovix® DS-11 FX+ , and Sanger sequenced in both the forward and reverse direction by submitting 6 ng of each product with 3.2 pmol of either primer in a total volume of 5 µl through the Genetic Analysis Service of the University of Otago (https://gas.otago.ac.nz). Forward and reverse sequences from each tissue sample were imported into Geneious Prime® v 2022.0.1 (Kearse et al. 2012). Sequences were checked for accuracy based on the electropherogram. Reverse sequences were reverse complemented, and a full barcode sequence was generated through pairwise alignment using the ‘Geneious Alignment’ with standard settings. The full barcode sequence was exported in.fasta format and imported into CRABS v 0.1.3 (Jeunen et al. 2022a) to generate a custom curated reference database (see “Bioinformatic analysis and taxonomy assignment” section for more information).
Laboratory processing of eDNA samples
Pre-PCR laboratory work was conducted in a designated PCR-free clean room. Prior to laboratory work, bench spaces and equipment were sterilized using a 10-min exposure to 10% bleach dilution (0.5% hypochlorite final concentration) and wiped with ultrapure water (UltraPure™ DNase/RNase‐Free Distilled Water, Invitrogen™) to reduce contamination risk (Prince and Andrus 1992). Additionally, negative control samples were processed alongside samples to investigate issues with cross-contamination. Negative control samples consisted of 50 µl ultrapure water for DNA extraction negatives and 2 µl ultrapure water for PCR no-template controls. Field controls were not collected on the fishing vessel due to extreme environmental conditions and sample collection being undertaken by fishery observers.
One tissue biopsy of ~ 0.5 cm3 was dissected from each sponge specimen for DNA extraction. DNA extraction followed the protocol described in (Jeunen et al. 2021). Briefly, DNA was extracted using the Qiagen DNeasy Blood and Tissue Kit according to the manufacturer’s recommendations, with slight modifications (Supplemental Table 3). DNA extracts were stored at − 20 °C until further processing.
Library preparation followed the protocol described in (Jeunen et al. 2018). Briefly, eDNA samples were analyzed for fish diversity using the fish (16S) metabarcoding assay (Berry et al. 2017), targeting a ~ 200 bp fragment of the 16S rDNA gene region. Prior to library preparation, input DNA for each sample was optimized using a dilution series (undiluted, tenfold dilution, 100-fold dilution) to identify inhibitors and low-template samples (Murray et al. 2015). Amplification was carried out in duplicate in 25 µl reactions. The qPCR mastermix consisted of 1 × SensiFAST SYBR Lo-ROX Mix (Bioline, London, UK), 0.4 µmol/l of each primer (Integrated DNA Technologies, Australia), 2 µl of template DNA, and ultrapure water as required. The thermal cycling profile included an initial denaturation step of 95 °C for 10 min; followed by 50 cycles of 30 s at 95 °C, 30 s at 54 °C, 45 s at 72 °C; and a final melt-curve analysis. A one-step amplification protocol using fusion primers was employed for library building (Berry et al. 2017). Fusion primers contained an Illumina adapter, a modified sequencing primer, a barcode tag (6–8 bp in length) and the template specific primer (Murray et al. 2015). Each sample was assigned a unique barcode combination (different forward and reverse barcodes). qPCR conditions followed the protocol as described above. Post qPCR, sample duplicates were pooled to reduce stochastic effects from PCR amplification (Alberdi et al. 2018; Leray and Knowlton 2015). Samples were then pooled into mini-pools based on end-point qPCR fluorescence, Ct-values, and melt-curve analysis (Murray et al. 2015). Size selection and qPCR clean-up followed the AMPure XP (Beckman Coulter, US) standard protocol. Mini-pools were visualized using gel electrophoresis to determine the presence of a single band and molarity of mini-pools was measured on Qubit. Pooling occurred equimolarly to produce a single DNA library. Due to differences in cycle number between samples and negative controls, the latter were spiked into the library to allow for optimal concentration of the library (Jeunen et al. 2019a). The resultant library was size selected once more using Pippin Prep (Cat # PIP0001; Sage Science, USA) and purified with Qiagen’s QIAquick PCR Purification Kit (Qiagen GmbH, Hilden, Germany) prior to final library quantitation on QIAxcel Advanced System (Qiagen GmbH, Hilden, Germany) and Qubit. Sequencing was performed on an Illumina MiSeq® using a 1 × 300 bp V2 Nano Illumina sequencing kit, following the manufacturer’s protocols, with 5% PhiX to minimize issues associated with low-complexity libraries.
Bioinformatic analysis and taxonomy assignment
Prior to the bioinformatic processing of sequencing data, raw fastq files were checked for quality using FastQC v 0.11.5 (Andrews 2010). Reads were demultiplexed and assigned to samples using cutadapt v 4.1 (Martin 2011), allowing for a single mismatch in the barcode and primer region. The assigned amplicons were filtered using ‘-fastq_filter’ function in USEARCH v 11.0.667 (Edgar 2010) based on a maximum expected error of 1.0 and minimum length of 150 bp. The success of quality filtering was checked in FastQC by comparing reports of FASTQ files before and after quality filtering. Remaining reads were dereplicated using the ‘-fastx_uniques’ function in USEARCH. Chimeric sequences were removed and ZOTUs (Zero-radius Operational Taxonomic Units) were generated using the ‘-unoise3’ function in USEARCH (Edgar 2016b). Finally, a ZOTU table was generated using the ‘-otutab’ function in USEARCH.
A custom curated reference database was generated using CRABS v 0.1.3 (Jeunen et al. 2022a). The custom curated reference database consisted of sequences downloaded from multiple online repositories ((‘db_download’ function) and in-house generated barcodes of Southern Ocean fish species (See “Fish reference barcodes” section; Supplemental File 1; ‘db_import’ function). Amplicon regions were extracted from sequences through in silico PCR analysis (‘insilico_pcr’ function) and pairwise global alignments (‘pga’ function). The ‘visualization’ function in CRABS was used to explore the reference database for missing barcodes (‘-method db_completeness’), mismatches in primer-binding regions (‘-method primer_efficiency’), and taxonomic resolution of the amplicon region (‘-method phylo’).
Taxonomy of ZOTUs were assigned using the ‘-sintax’ function in USEARCH (Edgar 2016a), with the custom curated reference databases generated by CRABS as input for the ‘-db’ parameter. ZOTUs were assigned to species level when a confidence of 1.00 was observed for the SINTAX algorithm and assigned to genus level for a confidence level between 0.97 and 0.99. The final taxonomic resolution of each taxonomic group was lowered to the lowest resolution observed between catch recordings and eDNA signals to enable accurate comparisons. For example, individuals within the Zoarcidae (eelpouts) family caught as bycatch are recorded as Zoarcidae, while CRABS identified no base pair mismatches in the amplicon regions for the genera Macrourus (rattails), Pogonophryne (barbeled plunderfishes), and certain species within the Trematomus genus (cod icefishes; CRABS ‘-method phylo’ function). After taxonomy assignment, the ZOTU table underwent final processing prior to statistical analysis, whereby (1) single read detections within each sample were removed to avoid issues related to tag jumping (Schnell et al. 2015), (2) reads were averaged between multiple sponges caught on a single line, and (3) the ZOTU table was transformed to relative abundance.
Statistical analysis and visualization
Rarefaction curves were generated from the unfiltered ZOTU table to assess sequencing coverage using the vegan v 2.5-7 package in R v 4.0.5 (R; http://www.R-project.org). Species richness was calculated for each sample and compared between eDNA and bycatch through multiple pairwise t-tests with Bonferroni correction to adjust for multiple comparisons using the rstatix v 0.7.0 package. Species accumulation curves were generated in the BiodiversityR v 2.13-1 package to assess differences in total number of fish species between eDNA and bycatch. Prior to beta diversity analyses, data tables were transformed to presence-absence. A permutational multivariate analysis of variance (PERMANOVA) was used to determine whether fish assemblage composition differed between eDNA and bycatch. Significant differences in dispersion between groups was tested (PERMDISP) to assess the reliability of PERMANOVA. A principal coordinate analysis (PCoA) was performed to visualize patterns of sample dissimilarity using the Jaccard index. Indicator values were calculated for each species using the labdsv v 2.0-1 package. Upper limits were set for indicator species, that is, species driving the difference in eDNA signal between sampling regions, to an indicator value index > 0.70 and a p value < 0.025 (Dufrêne and Legendre 1997). In this study, indicator values were used to determine the taxa driving the partitioning of samples between the two sampling regions found in the ordination analysis. Habitat preference of indicator species was used as biological validation of the difference found between sampling regions. Prior to Pearson correlation analysis, data tables were log-transformed to account for non-normal distributions. Pearson correlation was calculated using the ‘cor.test’ function within the native stats v 4.2.1 package. All bioinformatic and statistical scripts can be found in Supplemental Files 2 and 3.
Results
Fish catch recordings
Across all 26 fishing lines, a total weight of 22,834 kg of fish were recorded by fishery observers, with 16,479 kg caught on the continental slope and 6356 kg caught on the continental shelf (Supplemental Table 4a, b). Besides the target Antarctic toothfish, which constituted the highest abundance (22,151.5 kg; 97.0%) and count (955 individuals; 65.1%), an additional 14 fish species were recorded as bycatch. Taxonomic IDs within the Macrourus (Rattail fish) and Pogonophryne (Barbeled Plunderfish) genera were combined, as eDNA taxonomic resolution was set to genus level for both groups. Therefore, a total of 12 unique taxonomic IDs were caught by the fishing vessel, covering 9 families, 4 orders, and 2 classes. Macrourus sp. was the most by-caught taxon (1.3%), followed by the Antarctic starry skate (Amblyraja georgiana [Norman, 1938], Rajidae; 1.0%), and eel cod Muraenolepis sp. (0.3%).
Sequencing results
Demultiplexing of raw sequencing data resulted in assigning 156,854 sequences to eDNA samples (Supplemental Table 5). Filtering and quality control returned 144,671 (92.2%) sequences. Although PCR products of negative controls were spiked into the library, no sequences were returned after quality control. Denoising resulted in 27 ZOTUs, with 153,724 (98.0%) raw sequences matching to ZOTUs to create the unfiltered ZOTU table. After final quality filtration and taxonomy assignment, 153,695 (98.0%) sequences were incorporated for statistical analysis. Overall, eDNA samples achieved sufficient sequencing coverage based on the plateauing of rarefaction curves (Supplemental File 4) and mean number of reads per sample ± SD: 5123 ± 1443.
Taxonomy assignment returned 20 unique taxonomic IDs. After combining the taxonomic IDs of the Zoarcidae family (eelpouts), as bycatch recordings are limited to family level for this taxonomic group, a total of 18 unique taxonomic IDs were observed within eDNA samples, covering 11 families, 5 orders, and 2 classes (Supplemental Table 6). Overall, the Antarctic toothfish achieved the highest abundant eDNA signal (61.3%), followed by Macrourus sp. (15.0%), Chionobathyscus dewitti Andriashev & Neyelov, 1978 (Channichthyidae; 6.8%), and Trematomus sp. (6.5%).
Alpha and beta diversity comparison
A total of 20 fish taxa were detected across all sampling regions and monitoring methods, with a large overlap in species detection between methods across all samples and within each sampling region (Fig. 2; Table 1). While an eDNA signal for the skate genus, Bathyraja, was detected, we were unable to resolve the ZOTU to species level, despite the potential species-level taxonomic resolution reported during the in silico PCR analysis (Supplemental File 5). Thus, eDNA failed to distinguish two species recorded as bycatch, i.e., Eaton’s skate (Bathyraja eatonii [Günther, 1876]; Arhynchobatidae) and McCain’s skate (Bathyraja maccaini Springer, 1971; Arhynchobatidae).
Venn diagrams depicting species overlap between our eDNA survey and bycatch recordings for a all datapoints combined in purple, b the continental shelf sampling region in red, and c the continental slope sampling region in blue. Total number of species per monitoring method and proportion of species is represented between brackets. Venn diagram size is proportional to the number of detected species. Fish silhouette size is not representative of actual fish size. Number within silhouettes indicates species name as found in Supplemental Table 8
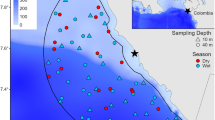
Overall, species were detected more frequently with eDNA than recorded as catch, except for Pogonophryne sp., Macrourus sp., and Dissostichus mawsoni, which were detected in the same number of samples (Fig. 3a). Significant differences in species richness were observed between monitoring methods within each sampling region according to multiple pairwise t-test with Bonferroni correction (continental shelf: t[8] = 8.4, p << 0.001; continental slope: t[16] = 3.9, p < 0.01), with eDNA significantly detecting a greater number of species on average compared to catch records (Fig. 3b). This result was further corroborated by species accumulation curves (Fig. 3c). Additionally, eDNA signals differed significantly in species richness between sampling regions according to Welch’s t-test (t[24] = 4.2, p < 0.001), with the continental slope containing a greater fish diversity over the continental shelf. Catch records, on the other hand, revealed no significant difference between sampling regions (t[24] = 0.7, p < 0.5).
a Frequency of taxon detection between eDNA and bycatch recordings. Maximum number of detections is 26. The red line (y = x) separates the taxa between those more frequently detected by eDNA (above) and those more frequently detected by catch (below). b Boxplots representing average species richness between eDNA and catch for the continental shelf (red) and continental slope (blue) sampling regions. Outliers are indicated by colored circles. The median is indicated by a black line within each boxplot. Significant differences, as indicated by multiple pairwise t-test with Bonferroni correction, are visualized with **p < 0.001 and ****p < 0.00005. c Species accumulation curves per sampling region (continental shelf [red] and continental slope [blue]) and monitoring method (eDNA [light color] and catch [dark color]). Number of samples are represented on x-axis and number of taxa on y-axis. The solid line indicates the average value, while shaded area depicts the standard error. d Principal Coordinates Analysis (PCoA) depicting similarity in community composition based on taxonomic incidence (Jaccard index; presence-absence), with the primary x-axis explaining 43.4% of the variation seen in the dataset and secondary y-axis explaining 15.5% of variation. Catch and eDNA data from the continental shelf are depicted in dark red triangles and light red crosses, respectively. Catch and eDNA data from the continental slope are depicted in dark blue circles and light blue plusses, respectively. Ellipses surrounding each group of samples represent 95% confidence intervals
Significant differences were also observed in community composition between sampling regions and monitoring methods according to PERMANOVA (sampling region: F1,48 = 86.6, p < 0.001; monitoring method: F1,48 = 19.6, p < 0.001), while no significant differences in dispersion were detected according to PERMDISP (F3,48 = 0.29; p < 1.0). PERMANOVA revealed sampling region (R2 = 0.54) to be the largest explanatory variable for the variation observed in the dataset, followed by monitoring method (R2 = 0.12). Community differences between sampling regions and monitoring methods were confirmed by ordination analysis (PCoA analysis; Jaccard index; presence-absence transformation; Fig. 3d), whereby sampling regions separated along the primary axis explaining 43.4% of the variation and monitoring methods separated along the secondary axis explaining 15.5% of the variation.
Due to differences in fish community detection between monitoring methods (alpha and beta diversity analyses), the indicator species analysis was conducted per monitoring method. The eDNA monitoring method identified two indicator species for the continental slope region, including Macrourus sp., and Muraenolepis sp., and five indicator species for the continental shelf region, including Blunt scalyhead (Trematomus eulepidotus Regan, 1914; Nototheniidae), Trematomus sp., Pogonophryne sp., Myers’ icefish (Chionodraco myersi DWitt & Tyler, 1960; Channichthyidae), and Jonah’s icefish (Neopagetopsis ionah Nybelin, 1947; Channichthyidae; Fig. 1b). Catch records identified the same two indicator species for the continental slope region, however, only identified a single indicator species for the continental shelf region, i.e., Trematomus sp. (Fig. 1b; Supplemental Table 7). Ecological descriptions from all indicator species identified in our dataset showed strong habitat preference in concordance with the spatial trend of the detections made either by eDNA or recorded as fish catch.
Catch abundance and biomass correlation to eDNA signal strength
Overall, eDNA signal strength and catch records showed significant correlation on a regional scale (Fig. 4). Across all samples, eDNA signal strength correlated better with catch abundance (R2 = 0.69; p < 0.005) compared to biomass (R2 = 0.41; p < 0.05). Additionally, correlation between eDNA and catch abundance was higher for the continental shelf region (R2 = 0.96; p << 0.001) compared to the continental slope region (R2 = 0.75; p < 0.005).
Correlation between relative eDNA signal strength and a catch abundance and b catch biomass. Linear regressions are shown for the full dataset (black; circles), the continental shelf sampling region (red; cross), and the continental slope sampling region (blue; triangle). Data was log–log transformed prior to analysis. Results are shown for all taxa detected using both methods
While regional fish diversity patterns correlated significantly between eDNA signal strength and catch abundance, single species correlation between monitoring methods displayed variable significance (Fig. 5). On average, low R2 values were obtained, indicating low predictive success for eDNA signal strength to estimate catch records, with the lowest R2 value of 0.0541 reported for biomass correlation of Zoarcidae and the highest R2 value of 0.8454 reported for abundance correlation of Trematomus sp. While the target fish, D. mawsoni, obtained a significant correlation between eDNA signal strength and catch abundance (p < 0.005), an R2 value of 0.32 was recorded due to the consistent number of specimens recorded and highly variable eDNA signal strength. Highly significant correlation and high R2 values were observed for Trematomus sp. (p << 0.001; R2 = 0.845), followed by Striped rockcod (Trematomus hansoni Boulenger, 1902; Nototheniidae; p << 0.001; R2 = 0.788), Macrourus sp. (p << 0.001; R2 = 0.523), and Muraenolepis sp. (p << 0.001; R2 = 0.493). Non-significant correlation was observed for Chionobathyscus dewitti (p < 0.1; R2 = 0.116) and Zoarcidae sp. (p < 0.1; R2 = 0.135).
Correlation between relative eDNA signal strength (x-axis) and catch biomass (blue; primary y-axis) and catch abundance (orange; secondary y-axis) for each taxon detected using both methods. Data was log–log transformed prior to analysis. Linear regression is indicated by a dashed line. The p-value, R2-value, and equation is provided above the graph for biomass and abundance for each species
Discussion
This study provides evidence for the application of using the naturally accumulated eDNA obtained from marine sponges (Cai et al. 2022; Jeunen et al. 2021; Mariani et al. 2019; Turon et al. 2020) caught as bycatch in demersal longlining fisheries to describe fish diversity patterns in the Ross Sea, Antarctica. Compared to fish catch records, eDNA metabarcoding allows for a more comprehensive investigation into fish biodiversity by detecting a larger proportion of the fish community. In addition, while our results show eDNA signal strength to be a suitable measurement for regional fish community composition (Salter et al. 2019; Stoeckle et al. 2021; Thomsen et al. 2016), the metabarcoding application is currently unable to predict catch numbers at the species level.
Our eDNA survey exceeded fish catch records in total number of fishes detected, as well as average species diversity per sample, thereby enabling us to gather additional biogeographical information on the data-limited Ross Sea ecosystem (Ainley 2002). This discrepancy between monitoring methods could have resulted from the high selectivity and bias of the hook-and-line fishing method endorsed by CCAMLR in the Ross Sea Antarctic toothfish fishery (Løkkeborg and Bjordal 1992; Moreno 1991). While selectivity and bias towards the target fish reduces bycatch, the utilization of such catch records to describe fish diversity patterns could potentially be hampered. Aquatic and natural sampler eDNA metabarcoding surveys, on the other hand, have previously been successfully implemented to describe biodiversity patterns in the marine environment (Berry et al. 2019; Jeunen et al. 2021; Nguyen et al. 2020; O’Donnell et al. 2017). Furthermore, aquatic eDNA metabarcoding surveys are frequently reported to outperform traditional monitoring surveys with regards to number of species detected (Afzali et al. 2020; Salter et al. 2019; Stat et al. 2019; Stoeckle et al. 2021; Thomsen et al. 2016). These previous findings agree with this study. The increase in number of species lies mainly in the manner of detection, whereby eDNA surveys do not rely on visual observations, but rather detect species indirectly through DNA released in the environment by inhabiting organisms, thereby increasing detection accuracy for low-abundant or elusive organisms (Mauvisseau et al. 2017; Simpfendorfer et al. 2016; Uthicke et al. 2022). Detection probability for eDNA surveys, however, might still be impacted through, e.g., (1) amplification bias (Kelly et al. 2019), (2) varying DNA shedding rates (Sassoubre et al. 2016; Wood et al. 2020), or (3) incomplete reference databases (Hestetun et al. 2020), potentially causing the reported false-negative detections in eDNA surveys in this study and elsewhere (Maiello et al. 2022; Stoeckle et al. 2021; Thomsen et al. 2016).
The only two species that could not be detected by our eDNA metabarcoding survey were the skates, Bathyraja eatonii and Bathyraja maccaini. While one eDNA signal for Bathyraja sp. was recovered, we were unable to resolve the ZOTU to species level. To date, 55 species have been described in this genus, with six species occurring in Antarctic and sub-Antarctic waters where they represent the dominant group of Chondrichthyan fauna (Long 1994; Smith et al. 2008). Only 12 species (21.8%) were represented with a reference barcode in our database, including three (50%) Antarctic and sub-Antarctic species, i.e., Bathyraja eatonii, Bathyraja maccaini, and Dark-belly skate (Bathyraja meridionalis Stehmann, 1987; Arhynchobatidae; Supplemental File 5). Given the multiple mismatches between the ZOTU and all reference barcodes and the species-level resolution of the amplicon region for the Bathyraja genus (based on the 12 available reference barcodes), it is likely the detected eDNA signal is from a described species lacking a reference barcode for the 16S rDNA gene or an undescribed species, as this taxonomic group is understudied with new species being described in recent years (Smith et al. 2008; Stehmann et al. 2021). Incomplete reference databases are, hence, a major limitation to eDNA surveys (Hestetun et al. 2020). It should be noted, however, that the reference barcodes used in this study for Bathyraja were obtained from the online data repository NCBI, known to contain erroneous sequences (Bagheri et al. 2020). Reference barcode validation could, therefore, lead to a reclassification to one of the two species recorded as bycatch. The reliance of eDNA to assign taxonomy based on online databases is powerful, because the entire community contributes to the completion of the reference database; but it is also a downside, due to lack of stringent curation potentially leading to misclassification (Bagheri et al. 2020). Additionally, only a single tissue biopsy was collected from each sponge specimens in our experiment. The inclusion of multiple replicate biopsies per sponge specimen could have increased the detection likelihood for rarer species due to the abundance distribution observed in eDNA metabarcoding data between high and low abundant DNA signals, thereby increasing the probability of detecting both skate species (Skelton et al. 2022). While eDNA most likely failed to detect both species, we cannot exclude the possibility of misidentification by fisheries observers, due to the understudied and poorly described nature of this taxonomic group (Smith et al. 2008; Stehmann and Bürkel 1990).
Highest taxonomic resolution was not consistently obtained for one survey method over the other. For example, taxonomic resolution of Macrourus sp., Pogonophryne sp., and Trematomus sp. had to be reduced to genus level for catch recordings, while taxonomic resolution of Zoarcidae sp. was reduced to family level for eDNA detection (Supplemental Table 8). While both monitoring methods achieved highest resolution for certain taxonomic groups, eDNA shows the highest potential for accurate, high-resolution taxonomy assignment by not relying on morphology-based identification (Seymour et al. 2021). To achieve this potential, reference databases will need to be completed and curated (Bagheri et al. 2020; Hestetun et al. 2020). Additionally, short amplicon primers with high taxonomic resolution and without amplification bias will need to be developed (Kelly et al. 2019). Alternatively, innovative sequencing technologies, such as Oxford Nanopore Technologies (ONT), enable longer DNA fragments to be sequenced, thereby potentially increasing the taxonomic resolution of eDNA metabarcoding data (Ames et al. 2021; Doorenspleet et al. 2021). Finally, the use of species-specific assays has the potential to increase the taxonomic resolution, as well as detection probability due to increased sensitivity over eDNA metabarcoding (Yu et al. 2022). However, the cost and time associated with the design of multiple specific assays is likely a hindrance for high diverse community monitoring (Yu et al. 2022).
Our natural sampler eDNA survey correlated highly to regional fish abundance and biomass observations from fish catch records, indicating quantitative information to be gathered from eDNA on a regional scale (Salter et al. 2019; Stoeckle et al. 2021; Thomsen et al. 2016). Similar observations were made for comparative experiments between aquatic eDNA and trawling (Salter et al. 2019; Stoeckle et al. 2021; Thomsen et al. 2016). However, the metric to which eDNA best correlated differed among studies, whereby Thomsen et al (2016) described variable results dependent on taxonomic resolution, Salter et al. (2019) found the highest correlation between eDNA and biomass, and Stoeckle et al (2021) identified an allometric index calculated from biomass to obtain the highest correlation. Further investigations into quantitative eDNA metabarcoding are, therefore, needed to tease out the current discrepancies between studies. While regional abundance information was obtained for our eDNA survey, we could not estimate fish catch numbers from a single line using eDNA metabarcoding. The lack of predictive power for our eDNA survey could be due to the bias of the longlining fishing method. For example, target species Dissostichus mawsoni biomass/abundance catch records were consistent across sampling regions, while eDNA signal strength varied. The consistent catch records could have been induced by the attraction of D. mawsoni to bait, thereby masking local abundance patterns (Kuriyama et al. 2018). For bycatch specimens, abundance/biomass estimates from catch records might be biased due to the selectivity of the gear against these organisms (Løkkeborg and Bjordal 1992). Hence, eDNA metabarcoding might be a better predictor of local fish abundance compared to longlining records. On the other hand, difficulties in obtaining abundance information from eDNA metabarcoding data are a well-known limitation of the methodology (Kelly et al. 2019). Environmental DNA signal strength can be influenced by biological (e.g., species-specific DNA shedding rates Kirtane et al. 2021), physical (e.g., environmental parameters Rourke et al. 2022), and technical (e.g., PCR amplification Kelly et al. 2019) factors, thereby potentially reducing the correlation between eDNA signal strength and taxon biomass/abundance.
Our results show the potential of using marine sponge bycatch specimens as a low-tech and cost-effective eDNA survey method to monitor the fish diversity in the Southern Ocean (Mariani et al. 2019). Marine sponges as natural eDNA samplers have been shown to achieve comparable results to aquatic eDNA surveys (Jeunen et al. 2021) and considered an innovative application of passive eDNA sampling (Bessey et al. 2021; Jeunen et al. 2022b), an effort to increase sample number in eDNA surveys by circumventing the need for the time-consuming step of active filtration (Bessey et al. 2021; Jeunen et al. 2022b). The use of marine sponge bycatch specimens in eDNA surveys will, furthermore, enable additional data gathering of the Porifera taxonomic group, which is among the less-studied benthic invertebrates with regards to extinction risk and conservation status (Bell et al. 2015). Additionally, yearly sponge bycatch sampling from the annual fishing season will enable us to investigate temporal biodiversity patterns associated with anthropogenic and climate impacts (Berry et al. 2019). Obtaining eDNA signals from previously collected sponge specimens stored in museums will also allow us to infer past ecosystem states and further our understanding of this understudied ecosystem to aid conservation efforts, such as the world’s largest marine protected area (Ballard et al. 2012).
Relying on bycatch specimens, however, could potentially hinder robust experimental design and consistent monitoring, as marine sponges are caught only when entangled on fishing lines dragging over the seafloor (Parker et al. 2009). For example, during the voyage underpinning this study, the San Aotea II fishing vessel recorded marine sponge specimens as bycatch in 26 (19.0%) out of the 137 fishing lines deployed. Additionally, the collection of specimens onboard commercial fishing vessels could potentially limit the opportunity for robust in-field contamination control, the standard approach within aquatic eDNA surveys (Takahashi et al. 2023). The upside of using bycatch specimens, on the other hand, is the lack of additional destructive sampling to gather data. An alternative consideration to marine sponges is the deployment of passive filtration devices, such as the metaprobe (Maiello et al. 2022) or artificial sponges (Jeunen et al. 2022b). Prior to deployment, however, it is imperative to investigate the impact of attached passive samplers to fishing lines to ensure commercial interests are not hindered. Autonomous sampling devices are another avenue currently being explored for eDNA surveys (Hansen et al. 2020; Yamahara et al. 2019). While initial success has been reported, deployment costs in remote areas, such as the Southern Ocean, and initial acquisition cost could hinder large-scale monitoring.
Conclusion
Effective conservation relies on detailed and extensive knowledge of the ecosystem. While international conservation goals have been put in place to limit the effect of detrimental anthropogenic pressures (Ballard et al. 2012), continuous monitoring is hindered by logistical constraints brought on from the remoteness of the Southern Ocean. In this experiment, we provide evidence for using marine sponge bycatch specimens as natural eDNA samplers to gain additional information on fish diversity patterns in the Southern Ocean. These passive samplers enabled us to survey a larger proportion of the fish community compared to fish catch records. Furthermore, proper curation of specimens and eDNA extracts will enable the re-examination of results when technological advances might allow for accurate abundance estimates and population genetic structure investigations. Finally, annual sponge bycatch collection and museum-stored sponges have the potential to let us uncover long-term temporal biodiversity patterns in relation to anthropogenic and climate impacts, thereby expanding our knowledge of this understudied ecosystem and aid conservation efforts.
Data availability
Raw and demultiplexed sequencing data have been deposited on Sequence Read Archive (SRA) and can be accessed through BioProject ID: PRJNA1022078. Bioinformatic and statistical scripts to analyze the demultiplexed sequencing data are made available in Supplemental File 2 and Supplemental File 3, respectively.
References
Afzali SF, Bourdages H, Laporte M, Mérot C, Normandeau E, Audet C, Bernatchez L (2020) Comparing environmental metabarcoding and trawling survey of demersal fish communities in the Gulf of St. Lawrence Canada. Environ DNA. https://doi.org/10.1002/edn3.111
Ainley DG (2002) The Ross Sea, Antarctica, where all ecosystem processes still remain for study, but maybe not for long. Mar Ornithol 30:55–62
Ainley DG, Pauly D (2014) Fishing down the food web of the Antarctic continental shelf and slope. Polar Rec 50(1):92–107. https://doi.org/10.1017/S0032247412000757
Alberdi A, Aizpurua O, Gilbert MTP, Bohmann K (2018) Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol Evol 9(1):134–147. https://doi.org/10.1111/2041-210X.12849
Allison EH, Perry AL, Badjeck M, Neil Adger W, Brown K, Conway D, Halls AS, Pilling GM, Reynolds JD, Andrew NL (2009) Vulnerability of national economies to the impacts of climate change on fisheries. Fish Fish 10(2):173–196
Ames CL, Ohdera AH, Colston SM, Collins AG, Fitt WK, Morandini AC, Erickson JS, Vora GJ (2021) Fieldable environmental DNA sequencing to assess jellyfish biodiversity in nearshore waters of the Florida keys, United States. Front Mar Sci. https://doi.org/10.3389/fmars.2021.640527
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge
Aronson RB, Thatje S, McClintock JB, Hughes KA (2011) Anthropogenic impacts on marine ecosystems in Antarctica. Ann N Y Acad Sci 1223(1):82–107
Bagheri H, Severin AJ, Rajan H (2020) Detecting and correcting misclassified sequences in the large-scale public databases. Bioinformatics (oxford, England) 36(18):4699–4705. https://doi.org/10.1093/bioinformatics/btaa586
Ballard G, Jongsomjit D, Veloz SD, Ainley DG (2012) Coexistence of mesopredators in an intact polar ocean ecosystem: the basis for defining a Ross Sea marine protected area. Biol Conserv 156:72–82. https://doi.org/10.1016/j.biocon.2011.11.017
Bell JJ, McGrath E, Biggerstaff A, Bates T, Cárdenas CA, Bennett H (2015) Global conservation status of sponges. Conserv Biol 29(1):42–53
Berry TE, Osterrieder SK, Murray DC, Coghlan ML, Richardson AJ, Grealy AK, Stat M, Bejder L, Bunce M (2017) DNA metabarcoding for diet analysis and biodiversity: a case study using the endangered Australian sea lion (Neophoca cinerea). Ecol Evol 7(14):5435–5453. https://doi.org/10.1002/ece3.3123
Berry TE, Saunders BJ, Coghlan ML, Stat M, Jarman S, Richardson AJ, Davies CH, Berry O, Harvey ES, Bunce M (2019) Marine environmental DNA biomonitoring reveals seasonal patterns in biodiversity and identifies ecosystem responses to anomalous climatic events. PLoS Genet 15(2):e1007943–e1007943. https://doi.org/10.1371/journal.pgen.1007943
Bessey C, Neil Jarman S, Simpson T, Miller H, Stewart T, Kenneth Keesing J, Berry O (2021) Passive eDNA collection enhances aquatic biodiversity analysis. Commun Biol 4(1):236. https://doi.org/10.1038/s42003-021-01760-8
Bowers HA, Pochon X, von Ammon U, Gemmell N, Stanton J-AL, Jeunen G-J, Sherman CDH, Zaiko A (2021) Towards the optimization of eDNA/eRNA sampling technologies for marine biosecurity surveillance. Water. https://doi.org/10.3390/w13081113
Cai W, Harper LR, Neave EF, Shum P, Craggs J, Arias MB, Riesgo A, Mariani S (2022) Environmental DNA persistence and fish detection in captive sponges. Mol Ecol Resour. https://doi.org/10.1111/1755-0998.13677
CCAMLR (2023) Scheme of international scientific observation scientific observer’s manual finfish fisheries
CCAMLR, S. committee (2022a) Compilation of member activity reports related to the Ross Sea region marine protected area, 2022
CCAMLR, S. committee (2022b) Korean Antarctic research and monitoring in the Ross Sea region in support of conservation measure 91–05
Clarke A, Johnston NM (2003) Antarctic marine benthic diversity. In: Oceanography and marine biology, an annual review, vol 41. CRC Press, Boca Raton, pp 55–57. ISBN: 9780429217715
Clarke A, Murphy EJ, Meredith MP, King JC, Peck LS, Barnes DKA, Smith RC (2006) Climate change and the marine ecosystem of the western Antarctic Peninsula. Philos Trans R Soc B Biol Sci 362(1477):149–166. https://doi.org/10.1098/rstb.2006.1958
Doorenspleet K, Jansen L, Oosterbroek S, Bos O, Kamermans P, Janse M, Wurz E, Murk A, Nijland R (2021) High resolution species detection: accurate long read eDNA metabarcoding of North Sea fish using Oxford nanopore sequencing. BioRxiv, 2021.11.26.470087. https://doi.org/10.1101/2021.11.26.470087
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67(3):345–366. https://doi.org/10.1890/0012-9615(1997)067[0345:SAAIST]2.0.CO;2
Eastman JT (2005) The nature of the diversity of Antarctic fishes. Polar Biol 28(2):93–107. https://doi.org/10.1007/s00300-004-0667-4
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics (oxford, England) 26(19):2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Edgar RC (2016a) SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. BioRxiv, 74161. https://doi.org/10.1101/074161
Edgar RC (2016b) UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 81257. https://doi.org/10.1101/081257
Fediajevaite J, Priestley V, Arnold R, Savolainen V (2021) Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol Evol 11(9):4803–4815. https://doi.org/10.1002/ece3.7382
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4(4):423 LP – 425
Fisheries New Zealand (2022) Aquatic environment and biodiversity annual review 2021
Griffiths HJ (2010) Antarctic marine biodiversity—what do we know about the distribution of life in the Southern Ocean? PLoS ONE 5(8):e11683
Hanchet S, Parker SJ, Mormede S (2015) Draft updated data collection plan for the Ross Sea toothfish fishery. https://meetings.ccamlr.org/en/wg-fsa-15/40
Hansen MC, Tolker-Nielsen T, Givskov M, Molin S (1998) Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol 26(2):141–149. https://doi.org/10.1111/j.1574-6941.1998.tb00500.x
Hansen BK, Jacobsen MW, Middelboe AL, Preston CM, Marin R, Bekkevold D, Knudsen SW, Møller PR, Nielsen EE (2020) Remote, autonomous real-time monitoring of environmental DNA from commercial fish. Sci Rep 10(1):13272. https://doi.org/10.1038/s41598-020-70206-8
Harper LR, Neave EF, Sellers GS, Cunnington AV, Arias MB, Craggs J, MacDonald B, Riesgo A, Mariani S (2023) Optimized DNA isolation from marine sponges for natural sampler DNA metabarcoding. Environ DNA. https://doi.org/10.1002/edn3.392
Hestetun JT, Bye-Ingebrigtsen E, Nilsson RH, Glover AG, Johansen P-O, Dahlgren TG (2020) Significant taxon sampling gaps in DNA databases limit the operational use of marine macrofauna metabarcoding. Mar Biodivers 50(5):70. https://doi.org/10.1007/s12526-020-01093-5
Jeunen GJ, Knapp M, Spencer HG, Taylor HR, Lamare MD, Stat M, Bunce M, Gemmell NJ (2018) Species-level biodiversity assessment using marine environmental DNA metabarcoding requires protocol optimization and standardization. Ecol Evol 9(3):1323–1335. https://doi.org/10.1002/ece3.4843
Jeunen GJ, Knapp M, Spencer HG, Lamare MD, Taylor HR, Stat M, Bunce M, Gemmell NJ (2019a) Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol Ecol Resour 19(2):426–438. https://doi.org/10.1111/1755-0998.12982
Jeunen GJ, Lamare MD, Knapp M, Spencer HG, Taylor HR, Stat M, Bunce M, Gemmell NJ (2019b) Water stratification in the marine biome restricts vertical environmental DNA (eDNA) signal dispersal. Environ DNA. https://doi.org/10.1002/edn3.49
Jeunen G-J, Urban L, Lewis R, Knapp M, Lamare M, Rayment W, Dawson S, Gemmell N (2020) Marine environmental DNA (eDNA) for biodiversity assessments: a one-to-one comparison between eDNA and baited remote underwater video (BRUV) surveys. https://doi.org/10.22541/au.160278512.26241559/v1
Jeunen G-J, Dowle E, Edgecombe J, von Ammon U, Gemmell N, Cross H (2022a) CRABS—a software program to generate curated reference databases for metabarcoding sequencing data. Mol Ecol Resour 23:725–738
Jeunen G-J, von Ammon U, Cross H, Ferreira S, Lamare M, Day R, Treece J, Pochon X, Zaiko A, Gemmell NJ, Stanton J-AL (2022b) Moving environmental DNA (eDNA) technologies from benchtop to the field using passive sampling and PDQeX extraction. Environ DNA. https://doi.org/10.1002/edn3.356
Jeunen G-J, Cane JS, Ferreira S, Strano F, von Ammon U, Cross H, Day R, Hesseltine S, Ellis K, Urban L (2021) Assessing the utility of marine filter feeders for environmental DNA (eDNA) biodiversity monitoring. BioRxiv
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics (oxford, England) 28(12):1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kelly RP, Shelton AO, Gallego R (2019) Understanding PCR processes to draw meaningful conclusions from environmental DNA studies. Sci Rep 9(1):12133. https://doi.org/10.1038/s41598-019-48546-x
Kirtane A, Wieczorek D, Noji T, Baskin L, Ober C, Plosica R, Chenoweth A, Lynch K, Sassoubre L (2021) Quantification of environmental DNA (eDNA) shedding and decay rates for three commercially harvested fish species and comparison between eDNA detection and trawl catches. Environ DNA 3(6):1142–1155. https://doi.org/10.1002/edn3.236
Koerich G, Fraser CI, Lee CK, Morgan FJ, Tonkin JD (2022) Forecasting the future of life in Antarctica. Trends Ecol Evol 38:24–34
Koziol A, Stat M, Simpson T, Jarman S, DiBattista JD, Harvey ES, Marnane M, McDonald J, Bunce M (2019) Environmental DNA metabarcoding studies are critically affected by substrate selection. Mol Ecol Resour 19(2):366–376. https://doi.org/10.1111/1755-0998.12971
Kuriyama PT, Branch TA, Hicks AC, Harms JH, Hamel OS (2018) Investigating three sources of bias in hook-and-line surveys: survey design, gear saturation, and multispecies interactions. Can J Fish Aquat Sci 76(2):192–207. https://doi.org/10.1139/cjfas-2017-0286
Leray M, Knowlton N (2015) DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proc Natl Acad Sci USA 112(7):2076–2081. https://doi.org/10.1073/pnas.1424997112
Løkkeborg S, Bjordal Å (1992) Species and size selectivity in longline fishing: a review. Fish Res 13(3):311–322. https://doi.org/10.1016/0165-7836(92)90084-7
Long DJ (1994) Quaternary colonization or paleogene persistence?: historical biogeography of skates (Chondrichtyes: Rajidae) in the Antarctic ichthyofauna. Paleobiology 20(2):215–228
Lynggaard C, Bertelsen MF, Jensen CV, Johnson MS, Frøslev TG, Olsen MT, Bohmann K (2022) Airborne environmental DNA for terrestrial vertebrate community monitoring. Curr Biol 32(3):701-707. e5. https://doi.org/10.1016/j.cub.2021.12.014
Maiello G, Talarico L, Carpentieri P, De Angelis F, Franceschini S, Harper LR, Neave EF, Rickards O, Sbrana A, Shum P, Veltre V, Mariani S, Russo T (2022) Little samplers, big fleet: eDNA metabarcoding from commercial trawlers enhances ocean monitoring. Fish Res 249:106259. https://doi.org/10.1016/j.fishres.2022.106259
Mariani S, Baillie C, Colosimo G, Riesgo A (2019) Sponges as natural environmental DNA samplers. Curr Biol 29(11):R401–R402. https://doi.org/10.1016/j.cub.2019.04.031
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17(1):10–12
Mauvisseau Q, Parrondo M, Fernández MP, García L, Martínez JL, García-Vázquez E, Borrell YJ (2017) On the way for detecting and quantifying elusive species in the sea: the Octopus vulgaris case study. Fish Res 191:41–48. https://doi.org/10.1016/j.fishres.2017.02.023
Moreno C (1991) Hook selectivity in the longline fishery of Dissostichus eleginoides (Nototheniidae) off the Chilean Coast. Sel Sci Pap (SC-CAMLR-SSP/8) 1991:107–118
Murray DC, Coghlan ML, Bunce M (2015) From benchtop to desktop: important considerations when designing amplicon sequencing workflows. PLoS ONE 10(4):1–21. https://doi.org/10.1371/journal.pone.0124671
Nguyen BN, Shen EW, Seemann J, Correa AMS, O’Donnell JL, Altieri AH, Knowlton N, Crandall KA, Egan SP, McMillan WO, Leray M (2020) Environmental DNA survey captures patterns of fish and invertebrate diversity across a tropical seascape. Sci Rep 10(1):6729. https://doi.org/10.1038/s41598-020-63565-9
O’Donnell JL, Kelly RP, Shelton AO, Samhouri JF, Lowell NC, Williams GD (2017) Spatial distribution of environmental DNA in a nearshore marine habitat. PeerJ 5:e3044. https://doi.org/10.7717/peerj.3044
Palumbi S (1991) Simple fool’s guide to PCR Version 2.0, privately published document compiled by S. Palumbi. Dept. Zoology, Univ. Hawaii, Honolulu, HI 96822
Parker SJ, Penney AJ, Clark MR (2009) Detection criteria for managing trawl impacts on vulnerable marine ecosystems in high seas fisheries of the South Pacific Ocean. Mar Ecol Prog Ser 397:309–317
Polanco Fernández A, Marques V, Fopp F, Juhel J-B, Borrero-Pérez GH, Cheutin M-C, Dejean T, González Corredor JD, Acosta-Chaparro A, Hocdé R, Eme D, Maire E, Spescha M, Valentini A, Manel S, Mouillot D, Albouy C, Pellissier L (2021) Comparing environmental DNA metabarcoding and underwater visual census to monitor tropical reef fishes. Environ DNA 3(1):142–156. https://doi.org/10.1002/edn3.140
Polanowski A, Clark J, Maschette D, Welsford DC, Deagle B (2018) Genetic identification of fish caught as by-catch in the Antarctic krill fishery and comparison with observer records. CCAMLR WG-EMM, WG-EMM
Prince AM, Andrus L (1992) PCR: how to kill unwanted DNA. Biotechniques 12(3):358–360
Rourke ML, Fowler AM, Hughes JM, Broadhurst MK, DiBattista JD, Fielder S, Wilkes Walburn J, Furlan EM (2022) Environmental DNA (eDNA) as a tool for assessing fish biomass: a review of approaches and future considerations for resource surveys. Environ DNA 4(1):9–33. https://doi.org/10.1002/edn3.185
Sala A, Azzali M, Russo A (2002) Krill of the Ross Sea: distribution, abundance and demography of Euphausia superba and Euphausia crystallorophias during the Italian Antarctic Expedition (January-February 2000). Sci Mar 66(2):123–133
Salter I, Joensen M, Kristiansen R, Steingrund P, Vestergaard P (2019) Environmental DNA concentrations are correlated with regional biomass of Atlantic cod in oceanic waters. Commun Biol 2(1):461. https://doi.org/10.1038/s42003-019-0696-8
Sassoubre LM, Yamahara KM, Gardner LD, Block BA, Boehm AB (2016) Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ Sci Technol 50(19):10456–10464. https://doi.org/10.1021/acs.est.6b03114
Schnell IB, Bohmann K, Gilbert MTP (2015) Tag jumps illuminated—reducing sequence-to-sample misidentifications in metabarcoding studies. Mol Ecol Resour 15(6):1289–1303. https://doi.org/10.1111/1755-0998.12402
Seymour M, Edwards FK, Cosby BJ, Bista I, Scarlett PM, Brailsford FL, Glanville HC, de Bruyn M, Carvalho GR, Creer S (2021) Environmental DNA provides higher resolution assessment of riverine biodiversity and ecosystem function via spatio-temporal nestedness and turnover partitioning. Commun Biol 4(1):512. https://doi.org/10.1038/s42003-021-02031-2
Simpfendorfer C, Kyne P, Noble T, Goldsbury J, Basiita R, Lindsay R, Shields A, Perry C, Jerry D (2016) Environmental DNA detects critically endangered largetooth sawfish in the wild. Endanger Species Res 30:109–116. https://doi.org/10.3354/esr00731
Skelton J, Cauvin A, Hunter ME (2022) Environmental DNA metabarcoding read numbers and their variability predict species abundance, but weakly in non-dominant species. Environ DNA. https://doi.org/10.1002/edn3.355
Smith PJ, Steinke D, Mcveagh SM, Stewart AL, Struthers CD, Roberts CD (2008) Molecular analysis of Southern Ocean skates (Bathyraja) reveals a new species of Antarctic skate. J Fish Biol 73(5):1170–1182. https://doi.org/10.1111/j.1095-8649.2008.01957.x
Smith WO, Ainley DG, Arrigo KR, Dinniman MS (2014) The oceanography and ecology of the Ross Sea. Ann Rev Mar Sci 6(1):469–487. https://doi.org/10.1146/annurev-marine-010213-135114
Stat M, John J, DiBattista JD, Newman SJ, Bunce M, Harvey ES (2019) Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity. Conserv Biol 33(1):196–205. https://doi.org/10.1111/cobi.13183
Stehmann MFW, Bürkel D (1990) Rajidae. In: Gon O, Heemstra PC (eds) Fishes of the Southern Ocean. J.L.B. Smith Institute of Ichthyology, Grahamstown, South Africa, pp 86–97. ISBN 0-86810-211-3
Stehmann MFW, Weigmann S, Naylor GJP (2021) First complete description of the dark-mouth skate Raja arctowskii Dollo, 1904 from Antarctic waters, assigned to the genus Bathyraja (Elasmobranchii, Rajiformes, Arhynchobatidae). Mar Biodivers 51(1):18. https://doi.org/10.1007/s12526-020-01124-1
Stevens DW, Dunn MR, Pinkerton MH, Forman JS (2014) Diet of Antarctic toothfish (Dissostichus mawsoni) from the continental slope and oceanic features of the Ross Sea region, Antarctica. Antarct Sci 26(5):502–512. https://doi.org/10.1017/S095410201300093X
Stoeckle MY, Adolf J, Charlop-Powers Z, Dunton KJ, Hinks G, VanMorter SM (2021) Trawl and eDNA assessment of marine fish diversity, seasonality, and relative abundance in coastal New Jersey, USA. ICES J Mar Sci 78(1):293–304. https://doi.org/10.1093/icesjms/fsaa225
Takahashi M, Saccò M, Kestel JH, Nester G, Campbell MA, van der Heyde M, Heydenrych MJ, Juszkiewicz DJ, Nevill P, Dawkins KL, Bessey C, Fernandes K, Miller H, Power M, Mousavi-Derazmahalleh M, Newton JP, White NE, Richards ZT, Allentoft ME (2023) Aquatic environmental DNA: A review of the macro-organismal biomonitoring revolution. Sci Total Environ 873:162322. https://doi.org/10.1016/j.scitotenv.2023.162322
Tejedo P, Benayas J, Cajiao D, Leung Y-F, De Filippo D, Liggett D (2022) What are the real environmental impacts of Antarctic tourism? Unveiling their importance through a comprehensive meta-analysis. J Environ Manag 308:114634. https://doi.org/10.1016/j.jenvman.2022.114634
Thomsen PF, Willerslev E (2015) Environmental DNA—an emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18. https://doi.org/10.1016/J.BIOCON.2014.11.019
Thomsen PF, Møller PR, Sigsgaard EE, Knudsen SW, Jørgensen OA, Willerslev E (2016) Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS ONE 11(11):e0165252
Trathan PN, Agnew D (2010) Climate change and the Antarctic marine ecosystem: an essay on management implications. Antarct Sci 22(4):387–398
Turon M, Angulo-Preckler C, Antich A, Præbel K, Wangensteen OS (2020) More than expected from old sponge samples: a natural sampler DNA metabarcoding assessment of marine fish diversity in Nha Trang Bay (Vietnam). Front Mar Sci 7:1042. https://doi.org/10.3389/fmars.2020.605148
Uthicke S, Robson B, Doyle JR, Logan M, Pratchett MS, Lamare M (2022) Developing an effective marine eDNA monitoring: eDNA detection at pre-outbreak densities of corallivorous seastar (Acanthaster cf. solaris). Sci Total Environ 851:158143. https://doi.org/10.1016/j.scitotenv.2022.158143
Verdier H, Konecny L, Marquette C, Reveron H, Tadier S, Grémillard L, Barthès A, Datry T, Bouchez A, Lefébure T (2021) Passive sampling of environmental DNA in aquatic environments using 3D-printed hydroxyapatite samplers. BioRxiv, 2021.05.12.443744. https://doi.org/10.1101/2021.05.12.443744
Weigand H, Beermann AJ, Čiampor F, Costa FO, Csabai Z, Duarte S, Geiger MF, Grabowski M, Rimet F, Rulik B, Strand M, Szucsich N, Weigand AM, Willassen E, Wyler SA, Bouchez A, Borja A, Čiamporová-Zaťovičová Z, Ferreira S, Dijkstra K-DB, Eisendle U, Freyhof J, Gadawski P, Graf W, Haegerbaeumer A, van der Hoorn BB, Japoshvili B, Keresztes L, Keskin E, Leese F, Macher JN, Mamos T, Paz G, Pešić V, Pfannkuchen DM, Pfannkuchen MA, Price BW, Rinkevich B, Teixeira MAL, Várbíró G, Ekrem T (2019) DNA barcode reference libraries for the monitoring of aquatic biota in Europe: gap-analysis and recommendations for future work. Sci Total Environ 678:499–524. https://doi.org/10.1016/j.scitotenv.2019.04.247
Wood SA, Biessy L, Latchford JL, Zaiko A, von Ammon U, Audrezet F, Cristescu ME, Pochon X (2020) Release and degradation of environmental DNA and RNA in a marine system. Sci Total Environ 704:135314. https://doi.org/10.1016/j.scitotenv.2019.135314
Xavier JC, Brandt A, Ropert-Coudert Y, Badhe R, Gutt J, Havermans C, Jones C, Costa ES, Lochte K, Schloss IR, Kennicutt MC, Sutherland WJ (2016) Future challenges in Southern Ocean ecology research. Front Mar Sci. https://doi.org/10.3389/fmars.2016.00094
Yamahara KM, Preston CM, Birch J, Walz K, Marin R, Jensen S, Pargett D, Roman B, Ussler W, Zhang Y, Ryan J, Hobson B, Kieft B, Raanan B, Goodwin KD, Chavez FP, Scholin C (2019) In situ autonomous acquisition and preservation of marine environmental DNA using an autonomous underwater vehicle. Front Mar Sci 6:373. https://doi.org/10.3389/fmars.2019.00373
Yu Z, Ito S, Wong MK-S, Yoshizawa S, Inoue J, Itoh S, Yukami R, Ishikawa K, Guo C, Ijichi M, Hyodo S (2022) Comparison of species-specific qPCR and metabarcoding methods to detect small pelagic fish distribution from open ocean environmental DNA. PLoS ONE 17(9):e0273670. https://doi.org/10.1371/journal.pone.0273670
Zhang S, Zhao J, Yao M (2020) A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish. Methods Ecol Evol 11(12):1609–1625. https://doi.org/10.1111/2041-210X.13485
Zhou S, Fan C, Xia H, Zhang J, Yang W, Ji D, Wang L, Chen L, Liu N (2022) Combined use of eDNA metabarcoding and bottom trawling for the assessment of fish biodiversity in the Zhoushan Sea. Front Mar Sci. https://doi.org/10.3389/fmars.2021.809703
Acknowledgements
We thank the onboard scientist Michael Prasad and the scientific observers Nigel Hollands (NZ/MPI observer) and Chuma Sijaji (CCAMLR international observer, Capricorn Fisheries) for their dedication of delivering scientific data and for carrying out sample collection. MPI project ANT201901 funded Michael Prasad being onboard of the San Aotea II fishing vessel. Marsden Fast-Start MFP-UOO2116 funded sample processing and sequencing costs. Ministry of Business, Innovation, and Employment Grant/Award Number MBIE ANTA1801 covered sample transport to and from fishing vessel and eDNA facilities at PML, University of Otago.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
The study design was conceptualized by GJJ, SM, SM, ML, and NJG. Sample collection was conducted by the FV San Aotea II fishing vessel. Laboratory work was performed by GJJ, JT, and SF. The bioinformatic analysis was conducted by GJJ. GJJ performed the statistical analysis, with input from SM, SM, ML, and NJG. GJJ wrote the manuscript with significant input from ML and NJG. All co-authors contributed to the writing of the manuscript and approve of the submission.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jeunen, GJ., Lamare, M., Devine, J. et al. Characterizing Antarctic fish assemblages using eDNA obtained from marine sponge bycatch specimens. Rev Fish Biol Fisheries 34, 221–238 (2024). https://doi.org/10.1007/s11160-023-09805-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11160-023-09805-3