Abstract
The pollination of self-incompatible diploid sweet cherry is determined by the S-locus alleles. We resolved the S-alleles of 50 sweet cherry cultivars grown in Estonia and determined their incompatibility groups, which were previously unknown for most of the tested cultivars. We used consensus primers SI-19/20, SI-31/32, PaConsI, and PaConsII followed by allele-specific primers and sequencing to identify sweet cherry S-genotypes. Surprisingly, 48% (24/50) of the tested cultivars, including 17 Estonian cultivars, carry the rare S-allele S17, which had initially been described in wild sweet cherries in Belgium and Germany. The S17-allele in Estonian cultivars could originate from ‘Leningradskaya tchernaya’ (S6|S17), which has been extensively used in Estonian sweet cherry breeding. Four studied cultivars carrying S17 are partly self-compatible, whereas the other 20 cultivars with S17 have not been reported to be self-compatible. The recommended pollinator of seven self-incompatible sweet cherries is of the same S-genotype, including four with S17-allele, suggesting heritable reduced effectiveness of self-infertility. We classified the newly genotyped sweet cherry cultivars into 15 known incompatibility groups, and we proposed four new incompatibility groups, 64–67, for S-locus genotypes S3|S17, S4|S17, S5|S17, and S6|S17, respectively, which makes them excellent pollinators all across Europe. Alternatively, the frequency of S17 might be underestimated in Eastern European populations and some currently unidentified sweet cherry S-alleles might potentially be S17.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sweet cherry (Prunus avium L.) is a fruit crop with growing popularity in Estonian home gardens. A wide range of local and foreign sweet cherry cultivars are grown, including 30 sweet cherry cultivars that have been bred in Estonia during the last half a century, primarily to improve their midwinter cold hardiness and increase the size and taste of the fruit (Jänes et al. 2010). Sweet cherry yield depends on planting appropriate pollinators nearby. Additionally, the right weather conditions, sufficient amount of pollen and corresponding blooming times are crucial factors for fertilization.
Most sweet cherry cultivars are self-incompatible, which is controlled by a single gametophytically expressed multiallelic S-locus (Crane and Lawrence 1928). In sweet cherry, the S-locus encodes two genes, the pollen-expressed F-box protein (SFB) and stylar ribonuclease (S-RNase) (Matsumoto and Tao 2016). Self-incompatibility occurs if both the pollen and the pistil carry the same S-allele (de Nettancourt 1977). Pollen is haploid and expresses SFB of one S-allele. Pistil is diploid and expresses S-RNases from both S-locus alleles. Additionally, S-locus F-box like proteins (SFLs) and S-haplotype-specific F-box proteins (SFBL), which are encoded by genes outside the S-locus, act as general inhibitors by recognising and mediating the polyubiquitination of S-RNases, thus keeping the environment in the pistil detoxified (Matsumoto and Tao 2016, 2019). When the pollen reaches pistil and its SFB-allele does not match either of the S-RNase alleles in the pistil, then S-RNases stay inactivated, and the pollen is able to fertilize the sweet cherry flower. However, when the SFB-allele in pollen matches either of the S-RNase alleles in the pistil, then pollen SFB inhibits polyubiquitination of S-RNases of the matching allele. Active S-RNase in the pistil causes a cytotoxic environment and disrupts pollen tube growth (Matsumoto and Tao 2016, 2019). Mutations, which cause absence of SFB in the pollen or truncated version of SFB, such as in alleles S3′, S4′, and S5′, result in self-fertility (Ushijima et al. 2004; Sonneveld et al. 2005; Marchese et al. 2007). Additionally, downregulation of MGST (M LOCUS-ENCODED GST) gene has also been shown to result in self-compatibility (Ono et al. 2018).
S-genotypes have been used to divide sweet cherry accessions into 63 incompatibility groups, one group of universal pollen donors, and one group of self-compatible cultivars (Schuster 2012, 2020; Marchese et al. 2017). Determination and assignment of the cultivars to the correct cross-compatibility groups are essential for sweet cherry breeding and crop production. Cross-compatibility groups and S-alleles have traditionally been identified by controlled pollination tests over several years and counting the number of formed fruits, which is very time and resource consuming and difficult to test. Molecular methods were developed to overcome these obstacles by being rapid, cheap and applicable regardless of the plants’ age and time of the year (Wiersma et al. 2001; Sonneveld et al. 2001, 2003). S-locus can also be used as a genetic marker for cultivar identification or pollen donor differentiation (Sebolt and Iezzoni 2009; Cachi and Wünsch 2014).
S-allele genotyping method is based on the knowledge that S-RNase gene alleles of sweet cherry have several conserved domains and two introns of varying size (Tao et al. 1999). Consensus primers for the polymerase chain reaction (PCR) have been designed to bind to the conserved regions of sweet cherry S-RNase so that they span either the first and/or the second intron (Tao et al. 1999; Wiersma et al. 2001; Sonneveld et al. 2001). Therefore, S-alleles are differentiated according to the size of the PCR-fragment. Consensus primer set SI-19 + SI-20 and SI-31 + SI-32 (Wiersma et al. 2001) can identify seven S-alleles (S1-S6 and S9). Another set of consensus primers, PaConsI-F + PaConsI-R and PaConsII-F + PaConsII-R (Sonneveld et al. 2003), can distinguish between 13 S-alleles (S1-S16). After S-RNase gene alleles were cloned, allele-specific PCR primers were developed (Sonneveld et al. 2001, 2003; Schuster et al. 2007; Szikriszt et al. 2012).
More than 30 S-alleles have been characterized both in cultivated and wild sweet cherries: S1-S16 (Sonneveld et al. 2001, 2003), S17-S22 (De Cuyper et al. 2005), S23-S25 (Wünsch and Hormaza 2004) and S27-S32 (Vaughan et al. 2008). However, the total number of unique S-alleles is currently 22 as several S-alleles have been found to be indistinct from each other, e.g. S8, S11 and S15 are not sufficiently different from S3, S7 and S5, respectively (Sonneveld et al. 2001, 2003; Schuster 2020). While some alleles are rarely found in cultivated sweet cherries (Schuster 2012), nearly all alleles are thought to originate from wild populations, with the possible exception of S5, which is very common in cultivated sweet cherries but seldom found in wild sweet cherries (Schuster 2012). Some S-alleles, such as S10, S13, S16, S17, S19, S21-S24, and S30, have been previously found mostly in wild sweet cherries and are rare in European cultivated sweet cherries (De Cuyper et al. 2005; Vaughan et al. 2008; Schuster 2012, 2020). Wild sweet cherry alleles S27-S29 and S31-S32 have not been found in cultivated sweet cherries thus far (Vaughan et al. 2008; Schuster 2020).
S-allele frequency varies widely in different regions (Lacis et al. 2008; Ipek et al. 2011; Schuster 2012; Cachi and Wünsch 2014; Ivanovych 2016). In Europe, S3, S6 and S1 are the most common S-alleles of sweet cherry cultivars (Schuster 2012). In the Baltic region, S3, S6 and S9 were the most frequent S-alleles found in Lithuanian sweet cherry cultivars (Stanys et al. 2008). A recent study of Russian sweet cherry cultivars detected S3, S5 and S6 as the most frequent S-alleles, but also suggested occurrence of rare S-alleles such as S17 and S30 or even yet undescribed S-alleles (Bezlepkina et al. 2020). However, in Latvian and Swedish sweet cherry collections only S-alleles S1-S6 were genotyped (Lacis et al. 2008). Estonia shares mainland border with Latvia and Russia and has partly similar climatic conditions; thus, sweet cherry S-allele frequency in Estonian sweet cherry cultivars could be similar to neighbouring countries. However, S-alleles of Estonian sweet cherries have not been verified thus far.
The objective of this study was to determine the S-locus alleles of sweet cherry cultivars grown in Estonia and to find the most suitable method for genotyping sweet cherry S-alleles for advancement of sweet cherry breeding in Estonia as well as to verify the list of sweet cherry recommended pollinators based on the S-locus genotyping.
Materials and Methods
Plant Materials and Genomic DNA Isolation
A total of 50 sweet cherry accessions widely grown in Estonia were analysed in this study, including 32 Estonian sweet cherry cultivars (n = 25) and breeding lines (n = 7). Samples were collected from Polli Horticultural Research Centre, Päidre Tree Nursery and from tissue culture collection in Estonian Crop Research Institute. Young sweet cherry branches were collected between March 2018 and December 2020. Branches were placed in water and forced to sprout. Genomic DNA was extracted from young leaves using a modified CTAB protocol. Plant tissue was ground in CTAB extraction buffer (2% cetyl trimethylammonium bromide w/v, 100 mM Tris–HCl, 1.4 M NaCl, 20 mM EDTA, 2% PVP-40 in water) and incubated for 25 min at 55 °C. The sample was centrifuged and supernatant pipetted into a new tube. Five hundred microliters of isopropanol was added and mixed by inverting several times. The mixture was incubated at –20 °C for 15 min, spun down at maximum speed, supernatant discarded and pellet was allowed to dry at room temperature. The pellet was dissolved in 50 µl of water. ‘Leningradskaya tchernaya’ is known and registered as ‘Leningradi must’ in Estonia.
S-Allele Genotyping
Estonian sweet cherries were S-genotyped using two methods: (1) consensus primers SI-19 + SI-20 and SI-31 + SI-32 (Wiersma et al. 2001), which can distinguish S1-S6 and S9 and (2) consensus primers PaConsI-F + PaConsI-R and PaConsII-F + PaConsII-R, which can distinguish S1-S16 (Sonneveld et al. 2003) (Table S1). Additionally, allele-specific primer pairs of sweet cherry S-alleles S1 to S17 were used for confirming genotypes (Sonneveld et al. 2001, 2003; Szikriszt et al. 2012) (Table S1). New S17-specific primers were also designed: PaS17_F3 (5′-GGGTCGCAATTTAAGAGATTGG-3′) and PaS17_R3 (5′-GCTGAGACATCCAAGCACATAG-3′) (Table S1).
PCR with SI-19 + SI-20 and SI-31 + SI-32 was optimized for FIREPol® Master Mix (Solis BioDyne, Estonia) with final concentrations: 1 × FIREPol® Master Mix with 2.5 mM MgCl2, 0.2 µM of each primer and 50 ng of genomic DNA. The reaction was performed as described by Wiersma et al. (2001), except annealing steps were performed at 54 °C for 90 s. Gel electrophoresis of SI-19 + SI-20 and SI-31 + SI-32 PCR products was run approximately for 2 h at 120 V using 1.5% agarose gel with ethidium bromide.
PCR with PaConsI-F + PaConsI-R and PaConsII-F + PaConsII-R primers was done as described by Sonneveld et al. (2003). FIREPol® DNA Polymerase (Solis BioDyne, Estonia) was used in all these PCR reactions. Overnight gel electrophoresis was performed at 50 V for 16 h using 2% agarose gel with ethidium bromide for PaConsI and 1.5% agarose gel for PaConsII PCR products.
Allele-specific PCR reactions were done as described by Sonneveld et al. (2001, 2003) and Szikriszt et al. (2012). Either FIREPol® DNA Polymerase (Solis BioDyne, Estonia) or DreamTaq DNA Polymerase (Thermo Scientific) (1 × DreamTaq Green buffer with 0.2 mM dNTP mix, 2 mM MgCl2, 0.2 µM of each primer and 50 ng of genomic DNA) was used in all these PCR reactions. PCR conditions for previously published S17-specific primers (EM-PC2consFD + PavS17R2) were optimized as following: 5 min at 94 °C, 40 cycles of 30 s at 94 °C, 30 s at 55 °C and 30 s at 72 °C, followed by 5 min at 72 °C. PCR conditions for new S17-specific primers were the following: 5 min at 94 °C, 35 cycles of 30 s at 94 °C, 30 s at 62 °C and 30 s at 72 °C, followed by 5 min at 72 °C. Gel electrophoresis was run at 90 V using 1.5% agarose gel with ethidium bromide.
To verify the presence or absence of S3′ self-compatible allele, primers by Sonneveld et al. (2005) were used to amplify and sequence SFB3 gene (Table S1). To verify the presence or absence of S4′ self-compatible allele, new primers to amplify 279 bp long fragment were designed for SFB4: SFB-4_F2 and SFB-4_R2 (Table S1). PCR conditions were 5 min at 95 °C, 40 cycles of 20 s at 95 °C, 20 s at 58 °C and 20 s at 72 °C, followed by 5 min at 72 °C. Gel electrophoresis was run at 90 V using 1.5% agarose gel with ethidium bromide. SFB4 PCR products were also sequenced.
Selected PCR products were Sanger sequenced at the University of Tartu, Institute of Genomics Core Facility with Applied Biosystems capillary electrophoresis analyser. Sequence alignment was conducted with Clustal Omega MSA tool by EMBL-EBI (Madeira et al. 2019). A selection of 32 sequences was also uploaded to the NCBI Nucleotide database and annotated with respective accession numbers (Table S2).
Assessment of Sweet Cherry Flowering Time
Sweet cherry flowers bloom before or during leaf budding. In Estonia, this usually occurs in the beginning or the first half of May. The beginning of flowering time was registered when 5% of flowers had bloomed. Full flowering (25% of flowers) starts 2–3 days later and lasted 6–10 days (25–75% of flowers). Flowering duration depends on the cultivar and the weather. On 2 years, also end of flowering was registered (when majority of petals have fallen).
Data Availability
Genomic DNA sequences have been submitted to NCBI Nucleotide database; accession numbers of the submitted sweet cherry genomic sequences are listed in Table S2.
Results
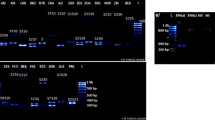
S-allele composition of 32 Estonian sweet cherry cultivars and 18 foreign cultivars grown in Estonia were analysed using two sets of consensus primers (Fig. 1 and Table 1). The first set of degenerate PCR primers (Wiersma et al. 2001) successfully identified the S-allele genotypes for 15 out of 50 studied cultivars. The second set of consensus primers PaConsI and PaConsII, which amplify introns I and II, respectively (Sonneveld et al. 2003), could determine S-allele genotypes for 23 out of 50 studied cultivars. S-allele composition was confirmed with allele-specific primer pairs (Sonneveld et al. 2001, 2003). Majority of the initially unidentified sweet cherry cultivars carried S-allele S17.
Agarose gel images of two sets of consensus primers for S-genotyping sweet cherry cultivars grown in Estonia. S-allele consensus primers SI-19 + SI-20 and SI-31 + SI-32 determine S-alleles S1-S6 and S9. S-allele consensus primers PaConsI-F + PaConsI-R and PaConsII-F + PaConsII-R determine S1-S16. S17 can be detected and is distinct from all other alleles with each consensus primer set. Agarose gel images of PCR products amplified by a SI-19 and SI-20 primers, b SI-31 and SI-32 primers, c PaConsI-F and PaConsI-R primers, and d PaConsII-F and PaConsII-R primers. Cultivars ‘Lapins’ S1|S4, ‘Kompaktnaja’ S2|S4, ‘Anne’ S3|S6, ‘Krupnoplodnaya’ S5|S9, ‘Madissoni roosa’ S4|S13 and ‘Irma’ S3|S17 were selected to represent all S-alleles detected in this study
High S17 Allele Frequency in Estonian Cultivars
S-genotyping of 32 Estonian sweet cherry cultivars (Table 1) resulted in detection of six different S-alleles (S3, S4, S5, S6, S13, S17), with S17 and S6 being the most frequent (Fig. 2). Among the 18 foreign and other cultivars grown in Estonia, we detected nine different S-alleles (S1, S2, S3, S4, S5, S6, S9, S13, S17), also with S17 being the most frequent. Altogether, 53% (17/32) of Estonian cultivars (e.g. ‘Piret’, ‘Polli rubiin’, ‘Tiia’) and 39% (7/18) of foreign cultivars (‘Aleksandrs’, ‘Dönisseni kollane’, ‘Elton’, ‘Gronkavaja’, ‘Leningradskaya tchernaya’, ‘Stella (False)’, ‘Zorka’) grown in Estonia carried the S-allele S17 (Table 1).
S-allele frequencies of 32 Estonian sweet cherry cultivars (blue bars) and 18 foreign sweet cherry cultivars (green bars) grown in Estonia in comparison to 545 European cultivars shown with gray bars in the background (Schuster 2012). The most common S-allele among both groups was S17
Neither of the consensus primer sets was initially able to clearly identify allele S17, although unidentified PCR products were detected. The unidentified PCR products were most similar to S5 with PaConsI primers and to S9 with PaConsII primers; however, neither was confirmed to be correct with allele-specific primers. The initially unidentified PCR product sizes using PaConsI and PaConsII were also similar to those previously reported for S17 (De Cuyper et al. 2005; Vaughan et al. 2008). Previously published allele-specific primers for S17 (Sutherland et al. 2004; Szikriszt et al. 2012) as well as newly designed more specific primers for S17 (PaS17_F3 + PaS17_R3) were used to confirm that indeed, S17 allele was the most frequent S-allele in Estonian sweet cherry cultivars (Table 1; Figs. 3 and S1). Thus, we confirmed that S17 can be detected with PaConsI and PaConsII. Moreover, allele S17 can be also detected, although less reliably, with SI-19 and SI-20 primers, where allele S17 sometimes has a weak band of about 750 bp and with SI-31 and SI-32 primers, where S17 had a band approximately 480 bp, which was indistinguishable from S1 and S5 allele (Fig. 1).
Comparison of two S17-specific primer pairs. New S17-specific primers amplify 398 bp long fragment specifically from the second intron of S-RNase of the S17 allele, whereas previously published S17-specific reverse primer combined with EM-PC2consFD amplifies also other S-locus alleles of different sizes. a Primers EM-PC2consFD (Sutherland et al. 2004) and PavS17R2 (Szikriszt et al. 2012) amplify S17 (505 bp), but also S3 (613 bp) and S4 (796 bp). b Primers PaS17_F3 and PaS17_R3 designed within this study amplify specifically only S17
The S-alleles of selected cultivars were sequenced in order to reverify the correct alleles. Sequencing confirmed that our S-genotyping results had been correct for alleles S3, S4, S6 and S17. Additionally, the presence of allele S16 was screened with allele-specific primers and was not detected.
S-allele S13 was found in five Estonian and two foreign cultivars. S13 can be detected using both consensus primer sets. Using SI-19 and SI-20 primers, alleles S9 and S13 may appear as S1 and S3 producing a band around 850 bp. For allele S13 primer pair SI-31 and SI-32 had a band of similar size as S4 and S6.
All samples of ‘Stella’ collected in Estonia were S-genotyped as S6|S17, although ‘Stella’ was previously reported to be S3|S4. An additional sample was requested from Latvia and S-genotyped as S3|S4’. Hence, we renamed the Estonian samples as ‘Stella (False)’ (Table 1).
New Incompatibility Groups Assigned
The results of S-allele genotyping of sweet cherry cultivars allowed the 50 cultivars to be classified into 19 known incompatibility groups including the proposed four new incompatibility groups, 64–67: LXIV, LXV, LXVI, and LXVII for S3S17, S4S17, S5S17 and S6S17, respectively (Tables S3 and S4).
Flowering Time Does Not Restrict Compatibility Between Cultivars
Pollination is only possible between cultivars flowering at the same time; hence, we analysed the empirical flowering time of sweet cherry cultivars grown in Estonia. Flowering time data from four different years (Fig. S2) showed that the flowering times of sweet cherry cultivars vary from year to year, but usually, the flowering times of different cultivars have an overlap. Some cultivars tend to flower earlier (e.g. ‘Ene’, ‘Taki’, ‘Tõmmu’) and some later (e.g. ‘Iput’, ‘Madissoni roosa’, ‘Zorka’, etc.); however, the early flowering cultivars are generally still in bloom when the late cultivars start to flower. Hence, the differences in flowering times are unlikely to cause incompatibility in pollination.
Known Self-Compatible Mutations Were Not Present in Estonian Cultivars
It has been reported that some cultivars grown in Estonia might be partly self-fertile, namely ‘Dönisseni kollane’ (S6|S17), ‘Elton’ (S6|S17), ‘Madissoni roosa’ (S4|S13), ‘Meelika’ (S4|S17) and ‘Veidenbergi maguskirss’ (S3|S17) (Table 2). Cultivar ‘Norri’ (S4|S17) is a recommended pollinator both for ‘Meelika’ (S4|S17) and ‘Mupi’ (S4|S17); however, all belong to the same incompatibility group (Table 2). Hence, sequencing analysis was used to detect possible clues for self-compatibility. Based on the sequence analysis, we showed that neither ‘Madissoni roosa’, ‘Meelika’, ‘Mupi’ nor ‘Norri’ have the S4′ deletion in the SFB4 gene (Fig. S3).
Another self-compatible mutation has been reported as S3′ haplotype, where complete deletion of SFB gene was detected (Sonneveld et al. 2005). Among sweet cherry cultivars grown in Estonia ‘Veidenbergi maguskirss’ (S3|S17) is the only partly self-fertile cultivar with S3 allele. Cultivar ‘Arthur’ (S3|S6) is a recommended pollinator both for ‘Polli murel’ (S3|S6) and ‘Anne’ (S3|S6); however, all belong to the same incompatibility group (Table 2). Additionally, cultivar ‘Tõmmu’ (S3|S17) is a recommended pollinator for ‘Polli rubiin’ (S3|S17) despite carrying identical pair of S alleles (Table 2). Hence, we analysed the presence or absence of SFB3 in the abovementioned cultivars grown in Estonia. We showed that all abovementioned cultivars carry SFB3 gene detected with primers PaSFB3-F and PaSFB3-R (Fig. S4). The sequence of the PCR fragments was identical to the published SFB3 sequence from the cultivar ‘Cristobalina’ (Fig. S4). Thus, the S3′ haplotype was not detected in the studied cultivars.
Discussion
Sweet cherry (Prunus avium L.) is a self-incompatible species, which requires a suitable cultivar to be planted in the vicinity for successful pollination and fruit-setting. The DNA locus responsible for sweet cherry self-incompatibility has been previously identified and genetic testing has been developed for determining S-locus alleles. We used two different sets of consensus primers (Wiersma et al. 2001; Sonneveld et al. 2003) together with allele-specific primers and sequencing to successfully identify all S-genotypes of 50 sweet cherry cultivars grown in Estonia. All genotypes were confirmed with allele-specific primer pairs (Sonneveld et al. 2001, 2003; Szikriszt et al. 2012; Table S5).
Estonian Sweet Cherry Cultivars Possess Rare S-Alleles
S-allele frequencies in Estonian sweet cherry cultivars were considerably different from allele frequencies reported among 545 European sweet cherry cultivars (Schuster 2012) (Fig. 2). The most striking difference is the high frequency of allele S17 in Estonia (present in 24 out of 50 cultivars). Allele S17 is extremely rare (allele frequency 0.01%) among European sweet cherry cultivars (Schuster 2012), but it is the most common S-allele in sweet cherries grown in Estonia, with allele frequencies of 26.6% and 19.4% in Estonian and foreign cultivars, respectively. Allele S17 was first described in Belgium, where 7 accessions with S17 were found among 65 wild sweet cherries (De Cuyper et al. 2005) and later as the third most common S-allele among 168 trees in a wild cherry population in Northern Germany (Schueler et al. 2006). It was found in a Sicilian sweet cherry cultivar ‘Moscatella’ (Marchese et al. 2007), in a Turkish wild sweet cherry accession (Coll.71) from among 28 landraces and wild sweet cherries (Szikriszt et al. 2012), in two landraces in a collection of 207 sweet cherries of diverse background (Mariette et al. 2010) and in three from among 94 cultivars mainly of Ukrainian origin (Ivanovych 2016). Allele S17 was listed among the rarest alleles with allele frequency of 0.006% among 153 Italian accessions (Marchese et al. 2017). Among cultivated sweet cherries, allele S17 is so rare that it has been reported in only six cultivars in a list of 1483 cultivars, including two Russian cultivars putatively with allele S17, and are usually designated to the incompatibility group 0 of universal donors (Schuster 2020). Four of these cultivars have S-genotype S6|S17. The notable spread of S17 among Estonian sweet cherry cultivars likely originates from Russian cultivar ‘Leningradskaya tchernaya’ S6|S17, and its offspring ‘Norri’ S4|S17, which have been extensively used in Estonian sweet cherry breeding. Estonian cultivar ‘Meelika’ S4|S17 was previously listed as S4|S6 (Lacis et al. 2008), but was now confirmed to be S4|S17. This suggests that there could be more cultivars with S17 that were originally genotyped to have more common alleles.
Alleles S1 and S2 are very common in Europe (Schuster 2012), but were found in none of the Estonian cultivars, and either of them were found only in one foreign cultivar genotyped in this study (‘Lapins’ S1|S4 and ‘Kompaktnaja’ S2|S4). Allele S3 is also less common in cultivars grown in Estonia than overall in Europe (Fig. 2). Allele S13, which is rare in Europe (2.0% allele frequency as reported by Schuster, 2012), was found in seven cultivars out of 50 (allele frequency 7.0%).
S-Genotyping Helps Differentiate Landraces and Old Cultivars
Sweet cherry cultivar ‘Dönisseni kollane’ S6|S17 appears distinct from ‘Dönissens gelbe knorpelkirsche’ S3|S6 (Schuster 2020). In addition to the difference in S-locus genotype, there is a potential difference in appearance as ‘Dönisseni kollane’ stone fruits have a slightly pink tone, which is visible in Estonian fruit catalogues but is not seen in pictures of ‘Dönissens gelbe knorpelkirsche’ in German fruit catalogues. Further research is needed to determine whether the Estonian 'Dönisseni kollane' is an offspring of ‘Dönissens gelbe knorpelkirsche’.
‘Elton’ is an old cultivar originating from England. According to the previously published genotyping, ‘Elton heart BC’ is S4|S6, ‘Elton heart EM’ is S3|S6 and ‘Black elton’ is S1|S3 (Wiersma et al. 2001; Wang et al. 2010; Schuster 2020). The ‘Elton’ clone grown in Estonia and used in this study was S-genotyped as S6|S17 (Table 1), matching with none of the previously published ‘Elton’ -named cultivars. We hypothesize that the ‘Elton’ grown and propagated in Estonia might be the progeny but not the direct clone of the ‘Elton' originating from England.
When comparing cultivars with their reported breeding parents (Table 2), the cultivars generally have one S-allele in common, which is the expected result for self-incompatible sweet cherry. An exception is ‘Zorka’ S13|S17, which shares neither of its S-alleles with its three reported offspring. Our S-genotyping results show that the sweet cherry collected as ‘Zorka’ S13|S17 is unlikely the breeding parent for ‘Polli murel’ S3|S6, ‘Anne’ S3|S6, and ‘Kaie’ S3|S6. ‘Zorka’ originates from Russian Federation bred in 1938 and is unable to survive very cold winters. A possible explanation could be that the original ‘Zorka’ has not survived and has been replaced by another cultivar of different S-genotype.
Comparing Empirical and Genotypical Cross-Compatibility of Sweet Cherry Cultivars
Crosspollination tests have been carried out in Estonia for a long period of time, and based on the accumulated empirical data (Jaama and Jaama 1992; Jänes 2016), a recommendation list of suitable pollinators has been created for sweet cherry cultivars grown in Estonia (Table 2). Many recommended cross-pollinating cultivars carried one allele in common with the cultivar being pollinated, meaning that it is generally sufficient for one allele to be different to achieve good pollination and yield. Intriguingly, in nine cases, the recommended cultivar had both alleles in common, thus belonging to the same incompatibility group; hence, its pollen should theoretically not be able to fertilize the pistil (Fig. 4 and Table 2).
Schematic overview of sweet cherry blossom and the role of S-alleles in self-incompatibility. a Pollination in self-incompatible sweet cherry can only occur when pollen is of a different S-allele than either of the alleles of the pistil. b Pollen with self allele triggers matching S-RNase to become cytotoxic and does not allow pollen tube to form. c Several sweet cherry cultivars grown in Estonia were found to be pollinated by (♀) or to pollinate (♂) other cultivars carrying the same S-alleles (Table 2). Some cultivars belong to both groups
For example, ‘Arthur’ and ‘Polli murel’ are both S3|S6 and at the same time recommended as suitable pollinators for each other. However, most cultivars with matching recommended pollinators have allele S17: ‘Elton’ S6|S17, ‘Kaspar’ S6|S17, ‘Norri’ S4|S17, ‘Polli rubiin’ S3|S17, ‘Meelika’, S4|S17 and ‘Mupi’ S4|S17 (Fig. 4c and Table 2). This implies that either these cultivars are partly self-fertile or there is more than one allele that appears as S17. We sequenced the PCR products of allele-specific primers of the cultivars with pollination discrepancies and confirmed our S-genotypes to be correct. We confirmed that the S-alleles of these cultivars are not different from each other; hence, the discrepancy must be due to self-fertility or possibly due to some environmental clue.
There exists a known self-fertile allele of S4′, which carries a deletion causing an early stop codon and degradation of SFB protein (Muñoz-Espinoza et al. 2017), which is found in ‘Stella’ and ‘Lapins’. Partly self-fertile ‘Meelika’ and ‘Madissoni roosa’ both have S4, which could be responsible for the self-fertility. Cultivars ‘Meelika’, ‘Mupi’ and ‘Norri’ were tested for the S4′ deletion but were found not to have the deletion (Fig. S3).
The cultivars with the ability to be pollinated by or to pollinate a cultivar of the same incompatibility group all carried alleles S3|S6 or S17. Out of five partly self-fertile cultivars, four have S17: ‘Dönisseni kollane’ S6|S17, ‘Elton’ S6|S17, ‘Meelika’ S4|S17 and ‘Veidenbergi maguskirss’ S3|S17 (Table 2). However, there are another 20 cultivars with S17, which are self-incompatible. ‘Leningradskaya tchernaya’ S6|S17, which has been extensively used as a breeding parent, is not officially a self-fertile sweet cherry but has occasionally been reported to be partly self-fertile in home gardens. ‘Leningradskaya tchernaya’ S6|S17 is also a breeding parent to partly self-fertile ‘Meelika’ S4|S17 . ‘Tiia’ S6|S17 is a curious case as it shares both of its S-alleles with its reported parent ‘Leningradskaya tchernaya’ S17, possibly confirming that ‘Leningradskaya tchernaya’ could have self-pollinated and produced a viable offspring. However, no cultivars have been found to be S17|S17; thus, it is not likely a self-compatible allele. The self-compatibility is also only partial, and yield is greatly improved when other cultivars are planted nearby. Thus, it could be a heritable reduced effectiveness of the gametophytic self-incompatibility but not a self-fertility allele in itself.
It is intriguing that several empirically identified recommended pollinators for Estonian sweet cherry cultivars belong to the same incompatibility group as the pollinated sweet cherry. Many of these discrepancies are with cultivars carrying S17, with four S17 cultivars being partly self-fertile. Cultivar ‘Madissoni roosa’ S4|S13 is also partly self-fertile but neither of the breeding parents are known. Hence, this possibly suggests a segregating mutation affecting self-compatibility in addition to the S-locus. The mechanism behind this is yet to be uncovered.
Sweet cherries grown in Estonia have very different S-allele distribution compared to the rest of Europe. Semi-wild S-allele S17 is vastly more common and S13 is fairly common, whereas S1 and S2 are not found in any Estonian cultivars or breeding lines. This makes Estonian cultivars excellent pollinators for sweet cherry all across Europe. Alternatively, the frequency of S17 could be underestimated in Eastern Europe populations, and this finding could elucidate S-genotypes for cultivars with previously unresolved S-genotype.
Availability of Data and Material
DNA sequences have been submitted to NCBI Nucleotide database. All relevant data has been included in the tables and supplementary tables of the manuscript.
References
Bezlepkina EV, Gulyaeva AA, Pikunova AV (2020) Aллeльный пoлимopфизм гeнa caмoнecoвмecтимocти y copтoв чepeшни ceлeкции BHИИCПК. Becтник Poccийcкoй Ceльcкoxoзяйcтвeннoй Hayки 26–28. https://doi.org/10.30850/vrsn/2020/4/26-28
Cachi AM, Wünsch A (2014) S-genotyping of sweet cherry varieties from Spain and S-locus diversity in Europe. Euphytica 197:229–236. https://doi.org/10.1007/s10681-014-1061-0
Crane MB, Lawrence WJC (1928) Genetical and cytological aspects of incompatibility and sterility in cultivated fruits. J Pomol Hortic Sci 7:276–301. https://doi.org/10.1080/03683621.1928.11513345
De Cuyper B, Sonneveld T, Tobutt KR (2005) Determining self-incompatibility genotypes in Belgian wild cherries. Mol Ecol 14:945–955. https://doi.org/10.1111/j.1365-294X.2005.02460.x
de Nettancourt D (1977) Incompatibility in angiosperms. Springer-Verlag, Berlin Heidelberg
Ipek A, Gulen H, Akcay ME et al (2011) Determination of self-incompatibility groups of sweet cherry genotypes from Turkey. Genet Mol Res GMR 10:253–260. https://doi.org/10.4238/vol10-1gmr1024
Ivanovych Y (2016) Sweet cherry genetic fingerprinting: methods and techniques. Marker-assisted selection (MAS) approaches for selection of sweet cherry varieties. https://doi.org/10.13140/RG.2.2.11739.98080
Jaama A, Jaama E (1992) Kirsid. Valgus
Jänes H (2016) Maalehe kirsiraamat: ahvatlevad kirsid koduaias. Hea lugu
Jänes H, Ardel P, Kahu K et al (2010) Some biological properties and fruit quality parameters of new sweet cherry cultivars and perspective selections. Agron Res 8:583–588
Lacis G, Kaufmane E, Rashal I et al (2008) Identification of self-incompatibility (S) alleles in Latvian and Swedish sweet cherry genetic resources collections by PCR based typing. Euphytica 160:155–163. https://doi.org/10.1007/s10681-007-9496-1
Madeira F, Park Y, mi, Lee J et al (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47:W636–W641. https://doi.org/10.1093/nar/gkz268
Marchese A, Giovannini D, Leone A et al (2017) S-genotype identification, genetic diversity and structure analysis of Italian sweet cherry germplasm. Tree Genet Genomes 13:93. https://doi.org/10.1007/s11295-017-1176-2
Marchese A, Tobutt KR, Raimondo A et al (2007) Morphological characteristics, microsatellite fingerprinting and determination of incompatibility genotypes of Sicilian sweet cherry cultivars. J Hortic Sci Biotechnol 82:41–48. https://doi.org/10.1080/14620316.2007.11512197
Mariette S, Tavaud M, Arunyawat U et al (2010) Population structure and genetic bottleneck in sweet cherry estimated with SSRs and the gametophytic self-incompatibility locus. BMC Genet 11:77. https://doi.org/10.1186/1471-2156-11-77
Matsumoto D, Tao R (2016) Distinct self-recognition in the Prunus S-RNase-based gametophytic Self-incompatibility System. Hortic J 85:289–305. https://doi.org/10.2503/hortj.MI-IR06
Matsumoto D, Tao R (2019) Recognition of S-RNases by an S locus F-box like protein and an S haplotype-specific F-box like protein in the Prunus-specific self-incompatibility system. Plant Mol Biol 100:367–378. https://doi.org/10.1007/s11103-019-00860-8
Muñoz-Espinoza C, Espinosa E, Bascuñán R et al (2017) Development of a molecular marker for self-compatible S4′ haplotype in sweet cherry (Prunus avium L.) using high-resolution melting. Plant Breed 136:987–993. https://doi.org/10.1111/pbr.12546
Ono K, Akagi T, Morimoto T et al (2018) Genome re-sequencing of diverse sweet cherry (Prunus avium) individuals reveals a modifier gene mutation conferring pollen-part self-compatibility. Plant Cell Physiol 59:1265–1275. https://doi.org/10.1093/pcp/pcy068
Schueler S, Tusch A, Scholz F (2006) Comparative analysis of the within-population genetic structure in wild cherry (Prunus avium L.) at the self-incompatibility locus and nuclear microsatellites. Mol Ecol 15:3231–3243. https://doi.org/10.1111/j.1365-294X.2006.03029.x
Schuster M (2012) Incompatible (S-) genotypes of sweet cherry cultivars (Prunus avium L.). Sci Hortic 148:59–73. https://doi.org/10.1016/j.scienta.2012.09.012
Schuster M (2020) Self-incompatibility (S) genotypes of cultivated sweet cherries -an overview update 2020. OpenAgrar Repos. https://doi.org/10.5073/20201016-141600
Schuster M, Flachowsky H, Köhler D (2007) Determination of self-incompatible genotypes in sweet cherry (Prunus avium L.) accessions and cultivars of the German Fruit Gene Bank and from private collections. Plant Breed 126:533–540. https://doi.org/10.1111/j.1439-0523.2007.01401.x
Sebolt A, Iezzoni A (2009) Utilization of the S-locus as a genetic marker in cherry to differentiate among different pollen donors. HortScience 44:1542–1546. https://doi.org/10.21273/HORTSCI.44.6.1542
Sonneveld T, Robbins TP, Bošković R, Tobutt KR (2001) Cloning of six cherry self-incompatibility alleles and development of allele-specific PCR detection. Theor Appl Genet 102:1046–1055. https://doi.org/10.1007/s001220000525
Sonneveld T, Tobutt KR, Robbins TP (2003) Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theor Appl Genet 107:1059–1070. https://doi.org/10.1007/s00122-003-1274-4
Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP (2005) Loss of pollen-S Function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype–specific F-box gene. Plant Cell 17:37–51. https://doi.org/10.1105/tpc.104.026963
Stanys V, Stanytė R, Stanienė G, Vinskienė J (2008) S-allele identification by PCR analysis in Lithuanian sweet cherries. Biologija 54:22–26. https://doi.org/10.2478/V10054-008-0005-9
Sutherland BG, Robbins TP, Tobutt KR, Weber WE (2004) Primers amplifying a range of Prunus S-alleles. Plant Breed 123:582–584. https://doi.org/10.1111/j.1439-0523.2004.01016.x
Szikriszt B, Doğan A, Ercisli S et al (2012) Molecular typing of the self-incompatibility locus of Turkish sweet cherry genotypes reflects phylogenetic relationships among cherries and other Prunus species. Tree Genet Genomes 9:155–165. https://doi.org/10.1007/s11295-012-0543-2
Tao R, Yamane H, Sugiura A et al (1999) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Am Soc Hortic Sci 124:224–233. https://doi.org/10.21273/JASHS.124.3.224
Ushijima K, Yamane H, Watari A et al (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J Cell Mol Biol 39:573–586. https://doi.org/10.1111/j.1365-313X.2004.02154.x
Vaughan SP, Bošković RI, Gisbert-Climent A et al (2008) Characterisation of novel S-alleles from cherry (Prunus avium L.). Tree Genet Genomes 4:531–541. https://doi.org/10.1007/s11295-007-0129-6
Wang H, Zhang K, Naihaoye Su H (2010) Identification of the S-genotypes of several sweet cherry (Prunus avium L.) cultivars by AS-PCR and pollination. Afr J Agric Res 5:250–256. https://doi.org/10.5897/AJAR09.491
Wiersma PA, Wu Z, Zhou L et al (2001) Identification of new self-incompatibility alleles in sweet cherry (Prunus avium L.) and clarification of incompatibility groups by PCR and sequencing analysis. Theor Appl Genet 102:700–708. https://doi.org/10.1007/s001220051700
Wünsch A, Hormaza JI (2004) Cloning and characterization of genomic DNA sequences of four self-incompatibility alleles in sweet cherry (Prunus avium L.). TAG Theor Appl Genet Theor Angew Genet 108:299–305. https://doi.org/10.1007/s00122-003-1418-6
Acknowledgements
We would like to thank Päidre Tree Nursery (Estonia) and Latvia University of Life Sciences and Technologies for samples of sweet cherries.
Funding
“National Programme for Plant Breeding from 2009–2019″ and „Collection and Conservation of Plant Genetic Resources for Food and Agriculture in 2014–2020”.
Author information
Authors and Affiliations
Contributions
K.L. conceived, initiated, and directed the study. A.K. designed the experiments and carried out the S-genotyping of sweet cherries. K.K. designed the genotype list, provided majority of the samples and provided empirical data about flowering and pollination. L.J. designed and carried out DNA sequence analyses and designed new allele-specific primers. A.K., K.L. and L.J. wrote the manuscript with revisions from K.K. All the authors contributed to and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
• Self-incompatibility allele S17 has previously been found in wild sweet cherries and only about a dozen cultivars. We report S17 allele in 24 previously unresolved sweet cherry cultivars. Four of the cultivars carrying S17 are partly self-fertile. New allele S17-specific primers were designed. We propose four new incompatibility groups.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kivistik, A., Jakobson, L., Kahu, K. et al. Wild and Rare Self-Incompatibility Allele S17 Found in 24 Sweet Cherry (Prunus avium L.) Cultivars. Plant Mol Biol Rep 40, 376–388 (2022). https://doi.org/10.1007/s11105-021-01327-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-021-01327-1