Abstract
Key message
S-RNase was demonstrated to be predominantly recognized by an S locus F-box-like protein and an S haplotype-specific F-box-like protein in compatible pollen tubes of sweet cherry.
Abstract
Self-incompatibility (SI) is a reproductive barrier that rejects self-pollen and inhibits self-fertilization to promote outcrossing. In Solanaceae and Rosaceae, S-RNase-based gametophytic SI (GSI) comprises S-RNase and F-box protein(s) as the pistil and pollen S determinants, respectively. Compatible pollen tubes are assumed to detoxify the internalized cytotoxic S-RNases to maintain growth. S-RNase detoxification is conducted by the Skp1-cullin1-F-box protein complex (SCF) formed by pollen S determinants, S locus F-box proteins (SLFs), in Solanaceae. In Prunus, the general inhibitor (GI), but not pollen S determinant S haplotype-specific F-box protein (SFB), is hypothesized to detoxify S-RNases. Recently, SLF-like proteins 1–3 (SLFL1–3) were suggested as GI candidates, although it is still possible that other proteins function predominantly in GI. To identify the other GI candidates, we isolated four other pollen-expressed SLFL and SFB-like (SFBL) proteins PavSLFL6, PavSLFL7A, PavSFBL1, and PavSFBL2 in sweet cherry. Binding assays with four PavS-RNases indicated that PavSFBL2 bound to PavS1, 6-RNase while the others bound to nothing. PavSFBL2 was confirmed to form an SCF complex in vitro. A co-immunoprecipitation assay using the recombinant PavS6-RNase as bait against pollen extracts and a mass spectrometry analysis identified the SCF complex components of PavSLFLs and PavSFBL2, M-locus-encoded glutathione S-transferase (MGST), DnaJ-like protein, and other minor proteins. These results suggest that SLFLs and SFBLs could act as predominant GIs in Prunus-specific S-RNase-based GSI.




Similar content being viewed by others
References
Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M et al (2018) The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46:W537–W544
Aguiar B, Vieira J, Cunha AE, Fonseca NA, Iezzoni A, Nocker S et al (2015) Convergent evolution at the gametophytic self-incompatibility system in Malus and Prunus. PLoS ONE 10:e0126138
Akagi T, Henry IM, Morimoto T, Tao R (2016) Insights into the Prunus-specific S-RNase-based self-incompatibility system from a genome-wide analysis of the evolutionary radiation of S locus-related F-box genes. Plant Cell Physiol 57:1281–1294
Babu V, Rajan V, D’Silva P (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics 9:433–446
Boivin N, Morse D, Cappadocia M (2014) Degradation of S-RNase in compatible pollen tubes of Solanum chacoense inferred by immunogold labeling. J Cell Sci 127:4123–4127
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Chen J, Wang P, de Graaf BHJ, Zhang H, Jiao H, Tang C et al (2018a) Phosphatidic acid counteracts S-RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell 30:1023–1039
Chen Q, Meng D, Gu Z, Li W, Yuan H, Duan X et al (2018b) SLFL genes participate in the ubiquitination and degradation reaction of S-RNase in self-compatible peach. Front Plant Sci 9:227
de Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants. Springer, Berlin
Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434
Dobriyal N, Tripathi P, Sarkar S, Tak Y, Verma AK, Sahi C (2017) Partial dispensability of Djp1’s J domain in peroxisomal protein import in Saccharomyces cerevisiae results from genetic redundancy with another class II J protein, Caj1. Cell Stress Chaperones 22:445–452
Entani T, Iwano M, Shiba H, Che FS, Isogai A, Takayama S (2003) Comparative analysis of the self-incompatibility (S) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8:203–213
Entani T, Kubo K, Isogai S, Fukao Y, Shirakawa M, Isogai A et al (2014) Ubiquitin–proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction. Plant J 78:1014–1021
Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S et al (2006) Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439:805–810
Hiratsuka S, Zhang S, Nakagawa E, Kawai Y (2001) Selective inhibition of the growth of incompatible pollen tubes by S-protein in the Japanese pear. Sex Plant Reprod 13:209–215
Igic B, Kohn JR (2001) Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA 98:13167–13171
Kakui H, Kato M, Ushijima K, Kitaguchi M, Kato S, Sassa H (2011) Sequence divergence and loss-of-function phenotypes of S locus F-box brothers (SFBB) genes are consistent with non-self recognition by multiple pollen determinants in self-incompatibility of Japanese pear (Pyrus pyrifolia). Plant J 68:1028–1038
Kubo K, Entani T, Tanaka A, Wang N, Fields AM, Hua Z et al (2010) Collaborative non-self recognition system in S-RNase-based self-incompatibility. Science 330:796–799
Kubo K, Paape T, Hatakeyama M, Entani T, Takara A, Kajihara K et al (2015) Gene duplication and genetic exchange drive the evolution of S-RNase-based self-incompatibility in Petunia. Nat Plant 1:14005
Kubo K, Tsukahara M, Fujii S, Murase K, Wada Y, Entani T et al (2016) Cullin1-P is an essential component of non-self recognition system in self-incompatibility in Petunia. Plant Cell Physiol 57:2403–2416
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li W, Chetlat RT (2014) The role of a pollen-expressed cullin1 protein in gametophytophytic self-incompatibility. Genetics 196:439–442
Li S, Sun P, Williams JS, Kao T (2014) Identification of the self-incompatibility locus F-box protein-containing complex in Petunia inflata. Plant Reprod 27:31–45
Li S, Williams JS, Sun P, Kao T (2016) All 17 S-locus F-box proteins of the S2- and S3-haplotypes of Petunia inflata are assembled into similar SCF complexes with a specific function in self-incompatibility. Plant J 87:606–616
Liu W, Fan J, Li J, Song Y, Li Q, Zhang Y et al (2014) SCFSLF-mediated cytosolic degradation of S-RNase is required for cross-pollen compatibility in S-RNase-based self-incompatibility in Petunia hybrida. Front Genet 5:228
Luu DT, Qin X, Laublin G, Yang Q, Morse D, Cappadocia M (2000) S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407:649–651
Matsumoto D, Tao R (2012) Isolation of pollen-expressed actin as a candidate protein interacting with S-RNase in Prunus avium L. J Jpn Soc Hortic Sci 81:41–47
Matsumoto D, Tao R (2016a) Distinct self-recognition in the Prunus S-RNase-based gametophytic self-incompatibility system. Hortic J 85:289–305
Matsumoto D, Tao R (2016b) Recognition of a wide-range of S-RNases by S locus F-box like 2, a general-inhibitor candidate in the Prunus-specific S-RNase-based self-incompatibility system. Plant Mol Biol 91:459–469
Muñoz-Sanz JV, Zuriaga E, Badenes ML, Romero C (2017) A disulfide bond A-like oxidoreductase is a strong candidate gene for self-incompatibility in apricot (Prunus armeniaca) pollen. J Exp Bot 68:5069–5078
Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y et al (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41
Niu S, Huang J, Zhang Y, Li P, Zhang G, Xu Q et al (2017) Lack of S-RNase based gametophytic self-incompatibility in orchids suggests that this system evolved after the monocot–eudicot split. Front Plant Sci 8:1106
Ono K, Akagi T, Morimoto T, Wünsch A, Tao R (2018) Genome re-sequencing of diverse sweet cherry (Prunus avium) individuals reveals a modifier gene mutation conferring pollen-part self-compatibility. Plant Cell Physiol 59:1265–1275
Pratas MI, Aguiar B, Vieira J, Nunes V, Teixeira V, Fonesca NA et al (2018) Inferences on specificity recognition at the Malus × domestica gametophytic self-incompatibility system. Sci Rep 8:1717
Qiao H, Wang H, Zhao L, Zhou J, Huang J, Zhang Y et al (2004) The F-box protein Ah SLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16:582–595
Ramanauskas K, Igić B (2017) The evolutionary history of plant T2/S-type ribonucleases. PeerJ 5:e3790
Sassa H, Kakui H, Miyamoto M, Suzuki Y, Hanada T, Ushijima K et al (2007) S locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175:1869–1881
Shirasawa K, Isuzugawa K, Ikenaga M, Saito Y, Yamamoto T, Hirakawa H, Isobe S (2017) The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res 24:499–508
Sijacic P, Wang X, Skirpan AL, Wang Y, Dowd PE, McCubbin AG et al (2004) Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429:302–305
Steinbachs JE, Holsinger KE (2002) S-RNase-mediated gametophytic self-incompatibility is ancestral in eudicots. Mol Biol Evol 19:825–829
Sun P, Kao T (2013) Self-incompatibility in Petunia inflata: The relationship between a self-incompatibility locus F-box protein and its non-self S-RNases. Plant Cell 25:470–485
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tao R, Yamane H, Sugiura A, Murayama H, Sassa H, Mori H (1999) Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J Amer Soc Hort Sci 124:224–233
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H (2003) Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15:771–781
Ushijima K, Yamane H, Watari A, Kakehi E, Hauck NR, Iezzoni AF et al (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39:573–586
Vieira J, Fonseca NA, Vieira CP (2008) An S-RNase-based gametophytic self-incompatibility system evolved only once in eudicots. J Mol Evol 67:179–190
Walsh P, Bursać D, Law YC, Cyr D, Lithgow T (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep 5:567–571
Williams JS, Der JP, dePamphilis CW, Kao T (2014a) Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflata. Plant Cell 26:2873–2888
Williams JS, Natale CA, Wang N, Li S, Brubaker TR, Sun P, Kao T (2014b) Four previously identified Petunia inflata S-locus F-box genes are involved in pollen specificity in self-incompatibility. Mol Plant 7:567–569
Zhao L, Huang J, Zhao Z, Li Q, Sim T, Xue Y (2010) The Skp1-like protein SSK1 is required for cross-pollen compatibility in S-RNase-based self-incompatibility. Plant J 62:52–63
Acknowledgements
This work was supported by a Grant-in-Aid (No. 16H06184) for Young Scientists (A) from the Japan Society for the Promotion of Science to D. M.
Author information
Authors and Affiliations
Contributions
D. M. designed and conducted the experiments and drafted the manuscript. Both authors edited the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2019_860_MOESM1_ESM.eps
Supplementary material 1 (EPS 1019 kb). Supplementary Fig. 1 Expression analysis of SLF-like and SFB-like homologs in Japanese apricot cv. Nanko pollen: expression levels were calculated from transcriptome data previously reported (DRR002283; Akagi et al. 2016) and expressed as FPKM
11103_2019_860_MOESM2_ESM.eps
Supplementary material 2 (EPS 1384 kb). Supplementary Fig. 2 The alignment of the N-terminal regions of the deduced amino-acid sequences of SLFL7 homologs of sweet cherry, peach and Japanese apricot:The MUSCLE program implemented in MEGA ver. 6.0 (Tamura et al. 2013) was used to generate the alignment. Amino acids shared over 75% among the homologs are shaded. The dashed line represents the region corresponding to the F-box motif in other Prunus SLFLs, SFB and SFBLs. Asterisks represent the amino-acid substitution in PavSLFL7A, which was shown to impair its interaction with PavSSK1
11103_2019_860_MOESM3_ESM.eps
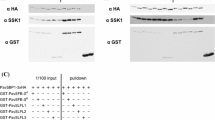
Supplementary material 3 (EPS 873 kb). Supplementary Fig. 3 The GST-pulldown assay of PavCul1A and Prunus avium Rbx1-like protein (XP_021828912.1). 3 × HA-tagged PavCul1A and GST-fused Rbx1-like protein were co-expressed in the cell-free system. The protein complexes bound to the glutathione sepharose beads were detected by the immunoblot
Rights and permissions
About this article
Cite this article
Matsumoto, D., Tao, R. Recognition of S-RNases by an S locus F-box like protein and an S haplotype-specific F-box like protein in the Prunus-specific self-incompatibility system. Plant Mol Biol 100, 367–378 (2019). https://doi.org/10.1007/s11103-019-00860-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00860-8