Abstract
Rice is a staple food for over half of the world’s population, but it contains only low amounts of bioavailable micronutrients for human nutrition. Consequently, micronutrient deficiency is a widespread health problem among people who depend primarily on rice as their staple food. Iron deficiency anemia is one of the most serious forms of malnutrition. Biofortification of rice grains for increased iron content is an effective strategy to reduce iron deficiency. Unlike other grass species, rice takes up iron as Fe(II) via the IRON REGULATED TRANSPORTER (IRT) in addition to Fe(III)-phytosiderophore chelates. We expressed Arabidopsis IRT1 (AtIRT1) under control of the Medicago sativa EARLY NODULIN 12B promoter in our previously developed high-iron NFP rice lines expressing NICOTIANAMINE SYNTHASE (AtNAS1) and FERRITIN. Transgenic rice lines expressing AtIRT1 alone had significant increases in iron and combined with NAS and FERRITIN increased iron to 9.6 µg/g DW in the polished grains that is 2.2-fold higher as compared to NFP lines. The grains of AtIRT1 lines also accumulated more copper and zinc but not manganese. Our results demonstrate that the concerted expression of AtIRT1, AtNAS1 and PvFERRITIN synergistically increases iron in both polished and unpolished rice grains. AtIRT1 is therefore a valuable transporter for iron biofortification programs when used in combination with other genes encoding iron transporters and/or storage proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron deficiency is the most common and widespread nutrient deficiency, particularly among children as well as women of childbearing age. Considering anemia as an indicator, 43 % of children, 38 % of pregnant woman and 29 % of non-pregnant women worldwide are estimated to be iron deficient (Steven et al. 2013). The main causes of iron deficiency include low bioavailable iron in the diet, increased requirements in pregnancy, or excessive blood loss caused by injuries, intestinal parasitic infection or menstruation. Increased dietary diversification, iron supplementation or food fortification are recommended interventions to overcome iron deficiency. Food fortification is considered the most effective and safest intervention in malaria-affected regions where iron supplementation leads to increased severity of the infectious disease (Oppenheimer et al. 1986; Sazawal et al. 2006). However, iron fortification of food is problematic since bioavailable iron compounds often react with other food components to cause off-flavors, color changes, or fat oxidation (Abbaspour et al. 2014). Agronomic practices, conventional breeding and/or genetic engineering are alternative approaches to enhance the nutritional content of staple crops. Rice is the main staple food for more than half of the world’s population and therefore an important target crop for biofortification. Rice is a poor source of dietary iron because the micronutrient-rich aleurone, bran and husk of the grain are removed during polishing. Most of the cultivated mega-rice varieties contain only around 2 µg iron per gram of grain after polishing (Bouis et al. 2011). Improving iron content in the rice endosperm using conventional breeding approaches alone has not been successful because the endosperm iron content has a very narrow range in the rice germplasm, as is the case for most other micronutrients (Slamet-Loedin et al. 2015). Therefore, genetic engineering is an important option for increasing iron content in rice endosperm (polished grains).
Genetic engineering approaches for iron biofortification reported to date have focused on overexpression of genes encoding the iron storage protein FERRITIN, iron transporters, and/or increasing production of iron chelating compounds. Endosperm-specific expression of FERRITIN, which can store up to 4500 Fe molecules in its central cavity, have increased iron content 2- to 3.7-fold in polished rice grains (Goto et al. 1999; Lucca et al. 2001; Oliva et al. 2014; Qu et al. 2005; Vasconcelos et al. 2003). Most graminaceous plants including rice utilize a chelation-based strategy for iron uptake and release phytosiderophores (PS) into the soil that bind ferric iron (Takagi 1976; Takagi et al. 1984). The PS-Fe(III) complexes are then transported into the root cells by specific transporters. All PS of the mugeneic acid (MA) family are synthesized from S-adenosyl-l-methionine via a conserved pathway of nicotianamine synthase (NAS), nicotianamine aminotransferase (NAAT) and deoxymugineic acid synthase (DMAS) (Kobayashi and Nishizawa 2012). Overexpression of the NAS gene resulted in a two- to four-fold increase in rice endosperm iron content (Masuda et al. 2009; Johnson et al. 2011; Lee et al. 2012). Other MA biosynthesis-related genes, for example, barley IRON DEFICIENCY-SPECIFIC 2 (IDS2), 3 (IDS3) and NAAT have also been introduced into rice, either alone or in combination with NAS (Kobayashi et al. 2008; Masuda et al. 2008; Mori et al. 2001; Suzuki et al. 2008; Takahashi et al. 2001) as well as with other iron transporters. Elevated iron levels were reported in transgenic rice lines overexpressing the combination of FERRITIN, NAS and RICE YELLOW STRIPE-LIKE 2 (OsYSL2) or of FERRITIN and NAS together with the barley NAAT and IDS3 genes (Aung et al. 2013; Masuda et al. 2012, 2013b). Several of the YELLOW STRIPE PROTEIN (YS) and YS-LIKE PROTEIN (YSL) family transporters have been evaluated in iron biofortification strategies. Transgenic expression of barley YS1 in rice increased leaf iron content by 1.5-fold, but no increase in grain iron was reported (Gomez-Galera et al. 2012). Constitutive expression of OsYSL15 resulted in a 1.2-fold increase of iron content in rice grains (Lee et al. 2009) and a phloem-specific overexpression of OsYSL2 increased iron content fourfold in rice endosperm (Ishimaru et al. 2010). Enhanced iron translocation from flag leaves to rice grains by reducing the expression of the vacuolar iron transport related genes OsVIT1 or OsVIT2 also resulted in a 1.8-fold increase of endosperm iron content (Bashir et al. 2013; Zhang et al. 2012). Additionally, constitutive overexpression of OsIRO2, which encodes an iron deficiency-inducible basic helix-loop-helix (bHLH) transcription factor regulating key genes involved in iron homeostasis increased iron content of the unpolished grains threefold when grown on calcareous soil (Ogo et al. 2011). The endosperm-specific expression of FERRITIN and PHYTASE combined with constitutive expression of NAS (NFP rice) gave a sixfold increase in the endosperm iron content in polished grains (Wirth et al. 2009). However, most of the iron biofortification strategies reported to date have not yet reached the target of 15 µg Fe/g (dry weight) in the rice endosperm (Bouis et al. 2011), except for the expression of a single OsNAS2 gene that produced lines with 14 and 19 µg/g iron in the polished grains (Johnson et al. 2011).
The IRON REGULATED METAL TRANSPORTER (IRT) is a member of the ZINC-REGULATED TRANSPORTER/IRON-REGULATED TRANSPORTER (ZRT/IRT1)-RELATED PROTEIN (ZIP) transporter family (Eng et al. 1998) and candidate for iron biofortification. In non-graminaceous plants, IRT facilitates Fe(II) uptake from the rhizosphere to the root cells (Eide et al. 1996; Guerinot 2000). Arabidopsis IRT1 is predominantly expressed in roots and upregulated in iron deficiency conditions (Eide et al. 1996). Unlike other graminaceous monocots that utilize chelation based strategy for Fe(III) uptake, rice can directly take up Fe(II) in water-logged paddy fields abundant in Fe(II) (Cheng et al. 2007). OsIRT1 and OsIRT2 encode Fe(II) transporters that are induced in iron deficient conditions (Ishimaru et al. 2006), confirming that rice has a functional Fe(II) uptake mechanism. We have introduced the Arabidopsis IRT1 gene into high-iron NFP rice lines (Wirth et al. 2009) under the control of pMsENOD12B, a Medicago sativa early nodulin gene promoter that is active in rice vascular tissue and root epidermal cells (Terada et al. 2001; Werthmüller 2000). NFP rice contains around 6 µg/gDW iron in polished grains when grown hydroponically and around 4 µg/g DW when grown in soil, in contrast to Taipei 309 control plants whose iron content in polished grains ranges between 1 and 2 µg/g DW. Expression of AtIRT1 in NFP lines (Wirth et al. 2009) significantly increases iron content in polished grains (9.6 µg/g DW) of soil grown plants, making AtIRT1 a strong candidate for iron biofortification of rice.
Results
MsENOD12B-controlled expression of AtIRT1 increases iron content in rice grains
AtIRT1 under the control of MsENOD12B promoter was transformed into Taipei 309 (TP309) and NFP rice (Wirth et al. 2009). Nine and twelve transgenic lines containing single transgene insertions in the TP309 background (IRT-TP309) and NFP background (IRT-NFP), respectively, were selected for growing the T1 generation. Based on the iron content in T2 polished grains (Supplementary Fig. 1), four IRT-TP309 (2, 3, 20 and 45) and IRT-NFP lines (6, 10, 36 and 37) each were selected for further analysis and were grown into T2 generation plants. T3 grains of IRT-TP309 and IRT-NFP lines had increased iron content in comparison to their non-transgenic controls (Fig. 1).
Iron content in the grains, leaves and roots of AtIRT1 expressing lines. Iron content in the T3 polished and unpolished grains, IRT-TP309 (a) and IRT-NFP (b). Iron content in the leaves of IRT-TP309 (c) and IRT-NFP (d) plants. Numbers indicate the selected lines. Iron content in the roots of IRT-TP309 (e) and IRT-NFP (f) plants. Values are the average of three biological replicates (±SD). Black and red asterisks above the bars indicate statistically higher and lower significant values calculated using Student’s T test, respectively, in comparison to the NTS or NFP controls (*P < 0.05; **P < 0.01). NTS non-transgenic sibling control
The iron content in polished grains of IRT-TP309 lines ranged from 3.78 to 4.86 µg/g DW as compared to 2.28 µg/g DW in non-transgenic sibling (NTS) control lines (Fig. 1a). Unpolished grains of IRT-TP309 lines had increased iron content ranging from 16.72 to 20.45 µg/g DW compared to 14.01 µg/g DW in NTS grains. The endosperm iron in line 3 was the highest with a 2.1-fold increase (4.86 µg/g) as compared to the control plants. Similarly, the IRT-NFP lines had up to 2.2-fold higher iron content (ranging from 6.38 to 9.67 µg/g DW) in polished grains as compared to the NFP lines (Fig. 1b). Unpolished grains of transgenic IRT-NFP lines contained 13.93 to 20.53 µg/g DW iron, which is variable as compared to the NFP control. These increase in iron content demonstrate the utility of the pMsENOD12B::AtIRT1 construct for iron biofortification in rice.
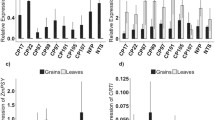
Leaf iron content of IRT-TP309 and IRT-NFP lines did not show significant differences to control plants, except IRT-NFP line 36 that had 1.2-fold higher leaf iron content (Fig. 1c, d). No significant change was observed in root iron content of IRT-TP309 lines, whereas three IRT-NFP lines had up to 2.3-fold increased iron as compared to NFP roots (Fig. 1e, f). However, the root iron content of these three IRT-NFP lines was not significantly different from TP309 (1068.12 µg/g DW iron) because of the reduced root iron content of the NFP siblings (Supplementary Fig. 2). Preliminary phenotypic characterization of T2 generation transgenic plants grown in soil showed variability for various scored parameters (days to flowering, plant height, SPAD value, one thousand grain weight; Supplementary Table 1). All transgenic lines had moderate but variable expression levels of AtIRT1 in both shoots and roots (Fig. 2), indicating that activity of the MsENOD12B promoter is not entirely restricted to rice vascular tissue and root epidermal cells as previously reported (Terada et al. 2001; Werthmüller 2000). IRT-TP309 line 45 had the highest AtIRT1 expression in the root and line 2 in the shoot, while IRT-NFP line 6 had high AtIRT1 expression levels in both shoot and root compared to the other IRT-NFP lines. The AtIRT1 expression differences among the independent transgenic lines did not correlate with the increased iron content observed in the grains, leaves and roots of these plants. Additionally, expression of several endogenous genes related to iron homeostasis was examined in selected transgenic lines. Although there were notable differences in the expression of the tested endogenous genes among the independent transformed lines, most genes showed a consistent expression (Supplementary Fig. 3; Supplementary Fig. 4).
Relative expression of transgene AtIRT1 in the independent transgenic lines. Relative AtIRT1 expression in root (white bar) and shoot (black bar) samples of 5-day-old seedlings in T3 generation is estimated. No expression of AtIRT1 was observed in NTS and NFP controls. The data were normalized with the endogenous expression of Os01g0147200 and Os11g0661400. Values are the average of three biological replicates (±SD)
Grains of AtIRT1 lines accumulate higher copper and zinc but not manganese
Among the metal ions copper, manganese and zinc were investigated in the T3 grains of AtIRT1 expressing lines. Copper was significantly increased in unpolished grains of both IRT-TP309 and IRT-NFP lines as compared to control lines (Fig. 3a, b). In the IRT-TP309 lines, the unpolished grains contained 8.56–9.80 µg/g DW copper (1.7-fold). However, only IRT-TP309 lines 2 and 20 showed a significant copper increase in polished grains with the highest copper content at 6.92 µg/g DW. Similarly, all four IRT-NFP lines had significant increases of copper in unpolished grains (up to 1.4-fold) while lines 10, 36 and 37 also had up to 1.8-fold copper increase in polished grains as compared to the NFP control (Fig. 3b). Furthermore, zinc content in polished grains was increased up to 1.5-fold in IRT-TP309 lines 3, 20 and 45 (Fig. 3c) and IRT-NFP line 36 (Fig. 3d) while IRT-NFP lines 6 and 37 showed decreased grain zinc content as compared to NFP plants (Fig. 3d). Similar increases of zinc content in the unpolished grains were observed for IRT-TP309 line 45 (1.3-fold) and IRT-NFP lines 10 and 36 (Fig. 3c, d). Unlike zinc and copper, the manganese content remained unchanged in grains of all the AtIRT1 expressing lines, except in IRT-TP309 line 3 in which manganese was slightly decreased (Fig. 3e, f).
Copper, zinc and manganese concentration in the grains of AtIRT1 expressing transgenic lines. T3 grains of the transgenic IRT-TP309 and IRT-NFP lines were analyzed for copper (a, b), zinc (c, d) and manganese content (e, f). Numbers indicate the selected lines. Values are the average of three biological replicates (±SD). Black and red asterisks above the bars indicate statistically higher and lower significant values calculated using Student’s T test, respectively, in comparison to the NTS or NFP controls (*P < 0.05; **P < 0.01)
Discussion
Iron biofortification of rice endosperm to dietary sufficient RDA levels has proven much more challenging compared to previously successful β-carotene biofortification of rice (Paine et al. 2005). This is likely due to the complex network of genes controlling iron homeostasis, including uptake, transport and storage, as well as iron transporters that can transport a broader range of metals (Thomine and Vert 2013). Efforts have been continuing for more than a decade to enhance iron levels in polished rice grains. Breeding for higher endosperm iron content is not possible because of the low endosperm iron variation in rice germplasm (Meng et al. 2005). Rice has therefore been transformed with genes encoding transporters mediating iron uptake and/or translocation, iron storage proteins and/or transcription factors controlling iron deficiency responses (Masuda et al. 2013a). Until recently, most transgenic iron biofortification approaches focused on phytosiderophore (PS) synthesis-related genes and the iron storage protein FERRITIN. Overexpression of NAS was a preferred approach because in addition to an increased production of PS in transgenic plants synthesis of nicotianamine (NA), a metal-chelator for internal iron transport (Stephan et al. 1996; Stephan and Scholz 1993), was also increased. Concerted expression of various gene combinations i.e., expressing genes related to uptake, translocation and storage from a single construct have generally been more successful than single transgene strategies. However, the transgene combinations might not always have a synergistic effect. Some of the studies also attempted increasing the sink in the rice endosperm by raising the FERRITIN expression levels, but with not much success. For example, overexpression of FERRITIN using both endosperm-specific GLOBULIN and GLUTELIN promoters did not increase iron content compared to expression of a single FERRITIN gene (Qu et al. 2005), most likely because of limiting iron mobilization from leaves during grain filling. Polished rice grains obtained from T2 transgenic lines overexpressing six transgenes (FERRITIN under the control of GLOBULIN and GLUTELIN promoters, barley NAAT-A, NAAT-B, NAS1 and IDS3), did not lead to further increases in iron content compared to dual FERRITIN overexpressing lines (Masuda et al. 2013b). Although different transgenic strategies have met varied levels of success to increase the iron content in both polished and unpolished rice grains, novel strategies are required to achieve further iron increases (at least 15 µg/g DW in polished grains) to meet the recommended dietary allowance levels.
The iron deficiency induced expression of OsIRT1 and OsIRT2, which are homologs of IRT1 in rice, encouraged the use of IRT in rice iron biofortification strategies (Lee and An 2009; Tan et al. 2015; Xiong et al. 2014). Transgenic rice expressing OsIRT1 under the control of the maize UBIQUITIN promoter were tolerant to iron deficiency and had over 1.1-fold increased iron content in the shoot and mature seeds but showed an aberrant growth phenotype in paddy fields (Lee and An 2009). The peanut IRT1 expressed under the control of an iron deficiency-inducible promoter in rice resulted in improved tolerance to low Fe availability in calcareous soils (Xiong et al. 2014). More recently, CaMV35S promoter-driven expression of apple IRT1 in rice resulted in increased iron content (up to 30.5 mg/kg) in mature seeds (Tan et al. 2015), but data on iron content in polished grains is not available from any of these studies. Thus, it is possible that a novel strategy of expressing an Fe(II) transporter such as IRT1 together with NAS and FERRITIN can further increase iron in the rice endosperm. To establish a proof of concept, we transformed the Arabidopsis IRT1 gene into our previously developed high-iron NFP rice (Wirth et al. 2009) in which NAS and FERRITIN genes synergistically increase grain iron content. To restrict the expression of AtIRT1 in transgenic rice we used the promoter of the early nodulin gene ENOD12B from alfalfa (Medicago sativa), which is expressed in nodules and also at low levels in roots, flowers, stems, and leaves (Bauer et al. 1994). Transgenic rice plants expressing a pMsENOD12B::GUS construct showed that the promoter is active in the vascular tissue as well as in epidermis and root hair cells (Terada et al. 2001; Werthmüller 2000). Our transgenic rice lines expressing pMsENOD12B::AtIRT1 in combination with NAS and FERRITIN genes had higher iron content in polished and unpolished grains as compared to the NFP plants. Together, our results show that restricted expression of AtIRT1 acts synergistically with NAS and FERRITIN to effectively increase iron in polished rice grains.
IRT1 has a high affinity for Fe(II) (Rogers et al. 2000) but also transports other divalent metal ions such as manganese, cadmium and zinc. Most of our AtIRT1 transgenic plants had increased grain copper content and in some lines, increased zinc content but no differences in manganese. However, these changes in the metal ion concentrations may not be a direct effect of AtIRT1 expression and the role of other metal homeostasis-related genes cannot be excluded. AtIRT1 expression may modulate the expression of other metal homeostasis related genes (Antosiewicz et al. 2014; Wang et al. 2013). We previously showed that several metal transport-related genes, including OsIRT1, OsZIP1, OsZIP3, OsZIP4, and OsYSL6 were upregulated in NFP lines (Wang et al. 2013). OsNAS3 and OsDMAS1, which encode enzymes in MA biosynthesis, were also upregulated in NFP rice when grown under low iron conditions (Wang et al. 2013). MA and NA also have a significant role in the transport of essential metals such as copper, iron, manganese and zinc (Haydon and Cobbett 2007). Therefore, similar modulation of metal homeostasis related endogenous genes could promote the uptake of other metal ions in addition to iron in the AtIRT1 transgenic rice lines. These possibilities can now be investigated in more detail using the IRT-TP309 and IRT-NFP lines. Together, restricted expression of AtIRT1 is an effective strategy for iron biofortification when used in combination with other iron transporters and/or iron storage proteins. In order to achieve further increases in iron, the biofortification strategies need to consider approaches going beyond the uptake of iron by the roots. It has become evident that even if increased amounts of iron can be transported into the plant, it does not necessarily reach the grain. Therefore, future strategies need to explore candidate genes that can promote effective translocation within the plant in combination with transporters that can increase availability of free iron for transport and storage into the grains.
Materials and methods
DNA construct, rice transformation and plant growth conditions
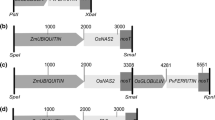
The construct expressing the Arabidopsis IRT1 gene [GenBank:U27590] under the control of MsENOD12B promoter was kindly provided by Dr. Christof Sautter (ETH Zurich). It was generated by amplifying AtIRT1 from a cDNA obtained from Arabidopsis roots using forward primer GGATCCTCTCATGAAAACAATCTT and reverse primer TCTAGAGCAGCAAAAGTTTTATTTATTT. The fragment was cut with BamHI and XbaI before inserting it into pMSB containing the MsENOD12B promoter (Terada et al. 2001). The transformation vector was transferred via Agrobacterium tumefaciens strain EHA105 (Hood et al. 1993) to Oryza sativa ssp. japonica cv. Taipei 309 (TP309) and to the previously developed high iron transgenic NFP rice (Wirth et al. 2009). Transformation, selection on hygromycin and regeneration were conducted as described by Nishimura et al. (2006). Plants were grown in a plant growth chamber with 60 % humidity/28 °C/16 h light and 8 h dark. Genomic DNA extraction was performed on leaves of 2-week-old plants using CTAB solution as described by Vasudevan et al. (2014). The presence of the transgene was verified by PCR using the primers AtIRT1fw, 5′-TGATGCTACCTTGAAGCTTAG-3′ and AtIRT1rv, 5′-TCAACTGCGCCGGAAGAATG-3′. Southern blot hybridization using digoxigenin (DIG) labeling was performed on BamHI digested genomic DNA of the transgenic lines to select for transformants with a single copy transgene insertion. The AtIRT1 fragment (889 base pairs) amplified by the AtIRT1 specific primers was used as a probe to detect the transgene. Example of Southern hybridization analysis is shown in Supplementary Fig. 5. The selected transformants, NFP and TP309 plants were grown in commercial soil (Klasmann-Deilmann GmbH, Germany) in the greenhouse conditions with 80 % humidity/30 °C/12 h light and 60 % humidity/22 °C/12 h dark. Mature grains were collected at 5 weeks after flowering for metal quantification. For metal quantification in the vegetative parts, shoot and root of 1-month-old seedlings of T3 generation were collected.
Metal ion measurements
Grain samples were de-husked to obtained unpolished brown grains. In order to obtain polished grains, the de-husked grains were processed with a grain polisher (Kett grain polisher ‘Pearlest’, Kett Electric Laboratory, Tokyo, Japan) for 1 min. Shoot and root samples were dried at 60 °C for 5 days. The samples were ground and 200 mg of each ground sample was boiled in 15 ml of 65 % v/v HNO3 solution at 120 °C for 90 min. Three ml of 30 % v/v H2O2 were subsequently added and continuously boiled at 120 °C for 90 min. Metal concentrations were determined using inductively coupled plasma-optical emission spectroscopy (ICP-OES) (Varian Vista-MPX CCD Simultaneous ICP-OES). The wavelength used for iron, zinc, manganese and copper were 238.204, 213.857, 257.610, and 324.754, respectively. The National Institute of Standards and Technology (NIST) rice flour standard 1658a was treated and analyzed in the same manner and used as quality control for every measurement. Data were analyzed using Student’s t test. The criteria of alpha = 0.05 and alpha = 0.01 was used to determine statistically significant differences among the tested lines.
Quantitative real-time PCR
Total RNA was extracted from root and shoot of 5-day-old seedlings in T3 generation using Trizol® reagent (Invitrogen, USA) and the RNA was treated with DNase I (Thermo Fisher Scientific Inc., USA) to remove genomic DNA contamination. First-strand cDNA was synthesized by RevertAid™ first strand cDNA synthesis kit (Thermo Fisher Scientific Inc., USA). Quantitative real-time PCR (qRT-PCR) was performed using Taqman hydrolysis probes (Roche, Switzerland) on 7500 FAST Real Time PCR system (Applied Biosystem, Inc., USA). Total reaction volume of 25 µl included 0.5 µl cDNA, 2.25 µl forward primer, 2.25 µl reverse primer, 0.25 µl probe (Roche Ltd., Switzerland), 12.5 µl Taqman® Gene Expression Mastermix (Applied Biosystems Ltd., USA) and 7.25 µl H2O. Primers were designed using Roche primer design website (https://lifescience.roche.com/shop/CategoryDisplay?catalogId=10001&tab=&identifier=Universal+Probe+Library&langId=-1&storeId=15006). Probe number and primer sequences are provided in Supplementary Table 2. The Ct value was obtained from 7500 Fast System Software (Applied Biosystems, Inc., USA). Primer efficiency was calculated by LinReg PCR (Tuomi et al. 2010). The data of qRT-PCR was normalized as described by Schefe et al. (2006).
References
Abbaspour N, Hurrell R, Kelishadi R (2014) Review on iron and its importance for human health. J Res Med Sci 19:164–174
Antosiewicz DM, Barabasz A, Siemianowski O (2014) Phenotypic and molecular consequences of overexpression of metal-homeostasis genes. Front Plant Sci 5:80. doi:10.3389/fpls.2014.00080
Aung MS, Masuda H, Kobayashi T, Nakanishi H, Yamakawa T, Nishizawa NK (2013) Iron biofortification of myanmar rice. Front Plant Sci 4:158. doi:10.3389/fpls.2013.00158
Bashir K, Takahashi R, Akhtar S, Ishimaru Y, Nakanishi H, Nishizawa NK (2013) The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice 6:31. doi:10.1186/1939-8433-6-31
Bauer P, Crespi MD, Szecsi J et al (1994) Alfalfa Enod12 genes are differentially regulated during nodule development by Nod factors and Rhizobium invasion. Plant Physiol 105:585–592
Bouis HE, Hotz C, McClafferty B, Meenakshi JV, Pfeiffer WH (2011) Biofortification: a new tool to reduce micronutrient malnutrition. Food Nutr Bull 32:S31–S40
Cheng L, Wang F, Shou H et al (2007) Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol 145:1647–1657. doi:10.1104/pp.107.107912
Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93:5624–5628
Eng BH, Guerinot ML, Eide D, Saier MH Jr (1998) Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J Membr Biol 166:1–7
Gomez-Galera S, Sudhakar D, Pelacho AM, Capell T, Christou P (2012) Constitutive expression of a barley Fe phytosiderophore transporter increases alkaline soil tolerance and results in iron partitioning between vegetative and storage tissues under stress. Plant Physiol Biochem 53:46–53. doi:10.1016/j.plaphy.2012.01.009
Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17:282–286. doi:10.1038/7029
Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465:190–198
Haydon MJ, Cobbett CS (2007) Transporters of ligands for essential metal ions in plants. New Phytol 174:499–506. doi:10.1111/j.1469-8137.2007.02051.x
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218
Ishimaru Y, Suzuki M, Tsukamoto T et al (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45:335–346. doi:10.1111/j.1365-313X.2005.02624.x
Ishimaru Y, Masuda H, Bashir K et al (2010) Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J 62:379–390. doi:10.1111/j.1365-313X.2010.04158.x
Johnson AA, Kyriacou B, Callahan DL, Carruthers L, Stangoulis J, Lombi E, Tester M (2011) Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS One 6:e24476. doi:10.1371/journal.pone.0024476
Kobayashi T, Nishizawa NK (2012) Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol 63:131–152. doi:10.1146/annurev-arplant-042811-105522
Kobayashi T, Nakanishi H, Takahashi M, Mori S, Nishizawa NK (2008) Generation and field trials of transgenic rice tolerant to iron deficiency. Rice 1:144–153
Lee S, An G (2009) Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ 32:408–416. doi:10.1111/j.1365-3040.2009.01935.x
Lee S, Chiecko JC, Kim SA et al (2009) Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol 150:786–800. doi:10.1104/pp.109.135418
Lee SC, Kim Y, Jeon U et al (2012) Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol Cells 33:269–275. doi:10.1007/s10059-012-2231-3
Lucca P, Hurrell R, Potrykus I (2001) Genetic engineering approaches to improve the bioavailability and the level of iron in rice grains. Theor Appl Genet 102:392–397. doi:10.1007/s001220051659
Masuda H, Suzuki M, Morikawa KC et al (2008) Increase in iron and zinc concentrations in rice grains via the introduction of barley genes involved in phytosiderophore synthesis. Rice 1:100–108
Masuda H, Usuda K, Kobayashi T et al (2009) Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2:155–166
Masuda H, Ishimaru Y, Aung MS et al (2012) Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci Rep 2:543. doi:10.1038/srep00543
Masuda H, Aung M, Nishizawa NK, Aung MS (2013a) Iron biofortification of rice using different transgenic approaches. Rice 6:19
Masuda H, Kobayashi T, Ishimaru Y et al (2013b) Iron-biofortification in rice by the introduction of three barley genes participated in mugineic acid biosynthesis with soybean ferritin gene. Front Plant Sci 4:132. doi:10.3389/fpls.2013.00132
Meng F, Wei Y, Yang X (2005) Iron content and bioavailability in rice. J Trace Elem Med Biol 18:333–338. doi:10.1016/j.jtemb.2005.02.008
Mori S, Nakanishi H, Takahashi M, Higuchi K, Nishizawa NK (2001) Genetic engineering of transgenic rice with barley strategy-II genes. In: Horst WJ (ed) Plant nutrition, vol 92. Developments in plant and soil sciences. Springer, Netherlands, pp 14–15. doi:10.1007/0-306-47624-X_5
Nishimura A, Aichi I, Matsuoka M (2006) A protocol for Agrobacterium-mediated transformation in rice. Nat Protoc 1:2796–2802. doi:10.1038/nprot.2006.469
Ogo Y, Itai RN, Kobayashi T, Aung MS, Nakanishi H, Nishizawa NK (2011) OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol Biol 75:593–605. doi:10.1007/s11103-011-9752-6
Oliva N, Chadha-Mohanty P, Poletti S et al (2014) Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol Breed 33:23–37. doi:10.1007/s11032-013-9931-z
Oppenheimer SJ, Gibson FD, Macfarlane SB, Moody JB, Harrison C, Spencer A, Bunari O (1986) Iron supplementation increases prevalence and effects of malaria: report on clinical studies in Papua New Guinea. Trans R Soc Trop Med Hyg 80:603–612
Paine JA, Shipton CA, Chaggar S et al (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotechnol 23:482–487. doi:10.1038/nbt1082
Qu le Q, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222:225–233. doi:10.1007/s00425-005-1530-8
Rogers EE, Eide DJ, Guerinot ML (2000) Altered selectivity in an Arabidopsis metal transporter. Proc Natl Acad Sci USA 97:12356–12360. doi:10.1073/pnas.210214197
Sazawal S, Black RE, Ramsan M et al (2006) Effects of routine prophylactic supplementation with iron and folic acid on admission to hospital and mortality in preschool children in a high malaria transmission setting: community-based, randomised, placebo-controlled trial. Lancet 367:133–143. doi:10.1016/S0140-6736(06)67962-2
Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H (2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J Mol Med (Berl) 84:901–910. doi:10.1007/s00109-006-0097-6
Slamet-Loedin IH, Johnson-Beebout SE, Impa S, Tsakirpaloglou N (2015) Enriching rice with Zn and Fe while minimizing Cd risk. Front Plant Sci 6:121
Stephan UW, Scholz G (1993) Nicotianamine: mediator of transport of iron and heavy metals in the phloem? Physiol Plant 88:522–529. doi:10.1111/j.1399-3054.1993.tb01367.x
Stephan U, Schmidke I, Stephan V, Scholz G (1996) The nicotianamine molecule is made-to-measure for complexation of metal micronutrients in plants. Biometals 9:84–90. doi:10.1007/BF00188095
Stevens GA, Finucane MM, De-Regi LM et al (2013) Global, regional, and national trends in haemoglobin concentration and prevalence of total and severe anaemia in children and pregnant and non-pregnant women for 1995–2011: a systematic analysis of population-representative data. Lancet Glob Health 1:e16–e25. doi:10.1016/S2214-109X(13)70001-9
Suzuki M, Morikawa KC, Nakanishi H, Takahashi M, Saigusa M, Mori S, Nishizawa NK (2008) Transgenic rice lines that include barley genes have increased tolerance to low iron availability in a calcareous paddy soil. Soil Sci Plant Nutr 54:77–85. doi:10.1111/j.1747-0765.2007.00205.x
Takagi S (1976) Naturally occurring iron-chelating compounds in oat-and rice-root washings: I. Activity measurement and preliminary characterization. Soil Sci Plant Nutr 22:423–433
Takagi S, Nomoto K, Takemoto T (1984) Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J Plant Nutr 7:469–477
Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S (2001) Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotechnol 19:466–469. doi:10.1038/88143
Tan S, Han R, Li P et al (2015) Over-expression of the MxIRT1 gene increases iron and zinc content in rice seeds. Transgenic Res 24:109–122. doi:10.1007/s11248-014-9822-z
Terada R, Ignacimuthu S, Bauer P et al (2001) Expression of early nodulin promoter gene in transgenic rice. Curr Sci 81:270–276
Thomine S, Vert G (2013) Iron transport in plants: better be safe than sorry. Curr Opin Plant Biol 16:322–327. doi:10.1016/j.pbi.2013.01.003
Tuomi JM, Voorbraak F, Jones DL, Ruijter JM (2010) Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50:313–322. doi:10.1016/j.ymeth.2010.02.003
Vasconcelos M, Datta K, Oliva N et al (2003) Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci 164:371–378. doi:10.1016/S0168-9452(02)00421-1
Vasudevan K, Vera Cruz CM, Gruissem W, Bhullar NK (2014) Large scale germplasm screening for identification of novel rice blast resistance sources. Front Plant Sci 5:505. doi:10.3389/fpls.2014.00505
Wang M, Gruissem W, Bhullar NK (2013) Nicotianamine synthase overexpression positively modulates iron homeostasis-related genes in high iron rice. Front Plant Sci 4:156. doi:10.3389/fpls.2013.00156
Werthmüller D (2000) Transgene expression in rice controlled by nodulation related legume promoters. Dissertation, ETH Zurich
Wirth J, Poletti S, Aeschlimann B et al (2009) Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol J 7:631–644. doi:10.1111/j.1467-7652.2009.00430.x
Xiong HC, Guo XT, Kobayashi T et al (2014) Expression of peanut Iron Regulated Transporter 1 in tobacco and rice plants confers improved iron nutrition. Plant Physiol Biochem 80:83–89. doi:10.1016/j.plaphy.2014.03.021
Zhang Y, Xu YH, Yi HY, Gong JM (2012) Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J 72:400–410. doi:10.1111/j.1365-313X.2012.05088.x
Acknowledgments
We gratefully acknowledge the generous support by Mrs. Jacqueline Imhof (Heinz Imhof Fellowship) and the ETH research grants for this work to WG and NKB. We thank Prof. Rainer Schulin (Soil protection group, ETH Zurich) for providing access to ICP-OES and Björn Studer for his kind assistance with metal measurements. We thank Irene Zurkirchen, Meng Wang, Kumar Vasudevan, Ting-Ying Wu and Isabel Janack for the technical support in the greenhouse and the laboratory. We thank Dr. Christof Sautter for providing the AtIRT1 expressing construct and the NFP rice used in this study.
Author contributions
NKB and WG designed the experiments, KB carried out the experiments, KB and NKB analyzed the data, KB and NKB wrote the manuscript, WG and NKB edited the manuscript. All authors have read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Boonyaves, K., Gruissem, W. & Bhullar, N.K. NOD promoter-controlled AtIRT1 expression functions synergistically with NAS and FERRITIN genes to increase iron in rice grains. Plant Mol Biol 90, 207–215 (2016). https://doi.org/10.1007/s11103-015-0404-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0404-0