Abstract
Biofortification of rice (Oryza sativa L.) using a transgenic approach to increase the amount of iron in the grain is proposed as a low-cost, reliable, and sustainable solution to help developing countries combat anemia. In this study, we generated and evaluated a large number of rice or soybean ferritin over-accumulators in rice mega-variety IR64, including marker-free events, by introducing soybean or rice ferritin genes into the endosperm for product development. Accumulation of the protein was confirmed by ELISA, in situ immunological detection, and Western blotting. As much as a 37- and 19-fold increase in the expression of ferritin gene in single and co-transformed plants, respectively, and a 3.4-fold increase in Fe content in the grain over the IR64 wild type was achieved using this approach. Agronomic characteristics of a total of 1,860 progenies from 58 IR64 single independent transgenic events and 768 progenies from 27 marker-free transgenic events were evaluated and most trait characteristics did not show a penalty. Grain quality evaluation of high-Fe IR64 transgenic events showed quality similar to that of the wild-type IR64. To understand the effect of transgenes on iron homeostasis, transcript analysis was conducted on a subset of genes involved in iron uptake and loading. Gene expression of the exogenous ferritin gene in grain correlates with protein accumulation and iron concentration. The expression of NAS2 and NAS3 metal transporters increased during the grain milky stage.
Similar content being viewed by others
Introduction
Deficiency of iron (Fe) affects about 40 % of the global population (World Bank 2006). Deficiency in dietary Fe is the principal cause of anemia, affecting more than 2 billion people worldwide, with women and children most at risk. Iron deficiency can increase the chances of maternal and child mortality due to severe anemia, and can have negative consequences for cognitive and physical development of children, and for physical performance (World Health Organization 2001, 2008). Combating micronutrient malnutrition is considered to be among the best investments to generate a high return in socioeconomic benefits (The Copenhagen Consensus 2004; www.copenhagenconsensus.com/).
Rice (Oryza sativa L.) is the most important staple food crop in the world, and it constitutes as high as 75 % of the daily calorie intake (Khush 2005). Unfortunately, rice grain contains low Fe and zinc (Zn) (Ghandilyan et al. 2006). Biofortification to increase Fe content in rice endosperm could provide a low-cost, sustainable strategy to remedy Fe deficiency in populations consuming polished rice as a staple food (Bajaj and Mohanty 2005; Zhao and Shewry 2011).
One strategy to increase Fe in rice endosperm is by incorporating exogenous ferritin genes through transgenic technology. Ferritin is a class of iron storage protein that can store up to 4,500 atoms of Fe per molecule in its central cavity (Theil 1987; Briat and Lobréaux 1997). Plant ferritin subunit sequences share 39 and 49 % identity with mammalian ferritin sequences (Briat et al. 2010). Ferritin sequesters excess Fe and protects cells against Fe-toxic effects (Theil 1987).
A ferritin-bioengineered rice diet was demonstrated to be as effective as a FeSO4 diet in replenishing hematocrit, hemoglobin concentration, and liver Fe concentrations using a hemoglobin repletion assay in rats (Beard et al. 1996; Murray-Kolb et al. 2002). Similarly, Lönnerdal (2009) reported that soybean ferritin is absorbed comparably to FeSO4 by in vitro assessment using human intestinal (CaCo-2) cells and in vivo by using radiolabeled ferritin in human subjects. Ferritin is as bioavailable as FeSO4 in non-anemic women, based on the study of Davila-Hicks et al. (2004).
Increased iron concentration by incorporation of soybean ferritin genes has been reported in rice (Goto et al. 1999; Vasconcelos et al. 2003; Qu et al. 2005), with a maximum increase of 3.7-fold in seed Fe concentration. Paul et al. (2012) transformed using a rice ferritin gene, OsFer2, with modest increase (twofold) in Fe concentration. Johnson et al. (2011) obtained a 4.4-fold increase using a single transgene chelator approach, nicotianamine synthase (OsNAS). Masuda et al. (2012) and Wirth et al. (2009) successfully applied multigene approaches, combining ferritin with a Fe transporter and a chelator, both reporting a sixfold increase in Fe levels in the grain. Wirth et al. (2009) obtained a final concentration of 6 mg kg−1and Masuda et al. (2012) reported a maximum increase of up to 7 mg kg−1 in the greenhouse; however, seed Fe concentrations were reduced to 4 mg kg−1 under field conditions. In maize, Drakaki et al. (2005) combined two transgenes (soybean ferritin and Aspergillus phytase) to increase bioavailable Fe in the endosperm. None of the above rice biofortification studies have utilized a large number of events in an elite mega-variety and intensively evaluated different rice and soybean ferritins under field conditions. Incorporation of a nutritional trait into an elite mega-variety possessing agronomic and quality traits preferred by farmers would have a prompt and wider impact (Manzanilla et al. 2011).
The use of selectable markers is of great importance for the selection of transformed cells in which foreign DNA integration has taken place because of the low efficiency of transgene integration. Studies have shown that one of the common antibiotic resistance genes [the hygromycin phosphotransferase (HPT) gene] used in the present study has no biosafety concerns (European Food Safety Authority Scientific Panel 2004; Goldstein et al. 2005; CERA 2010). In recent years, for better public acceptance, different techniques have been employed to generate marker-free transgenic plants, including the co-transformation approach (Miki and McHugh 2004), site-specific recombination (Kondrák et al. (2006), homologous recombination (Hare and Chua 2002), and non-selected transformation. In the co-transformation approach, transformation is achieved using two separate plasmid vectors: one containing the gene of interest and the other the selective marker gene that targets insertion at two different loci in the plant genome, to be further eliminated by progeny segregation.
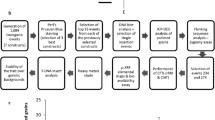
The objective of this study was to obtain high Fe concentration in polished rice that would be suitable for deregulation and adoption. A large number of transgenic plants containing the ferritin genes in the popular elite mega-variety IR64 was produced by the single gene approach in order to identify candidate events that have high Fe concentration in polished grain with good agronomic performance. This is the first study to perform extensive screening of a large population of progenies of transgenic rice events expressing the ferritin gene with and without the selectable marker under paddy soil conditions, suitable for easier deregulation as a potential transgenic product for rice farmers and consumers.
Materials and methods
Promoter selection
Three binary vectors were constructed to test the tissue-specific expression of rice endosperm-specific promoters, glutelin-1 (GLUB1, NCBI accession no. AY427569), glutelin-4 (GLUB4, NCBI accession no. AY427571), and globulin-1 (GLB1, NCBI accession no. AY427575), in IR64. The GLUB1 promoter sequence corresponds to the –12 to –2,336 position of the promoter sequence, the GLUB4 promoter sequence corresponds to the –12 to –1,474 position, and GLB1 corresponds to the –1 to –840 position of the promoter sequence. The GLUB1 promoter sequence was amplified using rice genomic DNA of Nipponbare using forward primer 5′-ACAGATTCTTGCTACCAACA-3′ and reverse primer 5′-ACGGATCCCCTTGCTTATGGAAACTTAAG-3′. The GLUB4 promoter sequence was amplified using rice genomic DNA using forward primer 5′-TACAGGGTTCCTTGCGTGAA-3′ and reverse primer 5′-ACGGATCC ATGTTATTGGAAACTTGGGC-3′. GLB1 was amplified using forward primer 5′-GTTAATCATGGTGTAGGCAA-3′ and reverse primer 5′-ACGGATCCGGTTGTTGTAGGACTAATGAAC-3′. The promoter fragments were cloned separately in pCR 2.1 TOPO (Invitrogen, San Diego, CA, USA), namely as pTOPO–GLUB1, pTOPO–GLUB4 and pTOPO–GLB1.
These promoters were fused individually to the GUSA reporter gene in the binary vector pCAMBIA 1381Z (NCBI accession no. AF234306). The resulting vectors were transferred into Agrobacterium strain LBA4404 by the freeze–thaw method. These Agrobacterium strains harboring the different vectors individually were used to infect immature embryo from immature seeds of Oryza sativa cv IR64.
A histochemical β-glucuronidase (GUS) assay was performed as described by Jefferson et al. (1987) for dough stage [15 days after flowering (DAF)] and mature (30 DAF) seeds in selected transgenic lines and wild-type IR64. Reverse-transcriptase (RT) PCR using virA primers was performed to determine whether GUS expression due to Agrobacterium contamination was present in the tissue samples.
Construct design of transformation vectors
Ferritin cDNAs were isolated from soybean (Glycine max L. cv PHI29924) and rice (O. sativa cv Nipponbare). Total mRNA was isolated from 1 g (fresh weight) of leaf tissues and the ferritin cDNAs SoyFERH1 (0.914 kb), SoyFERH2 (0.803 kb), OsFER1C (0.820 kb), and OsFER2C (0.821 kb) were amplified by RT-PCR using forward primer 5′-ACGTCGACCACAAATCTTAGCCGCCATT-3′ and reverse primer 5′-ACCTGCAGCCAGAATTTCAGAAAAGACCAAATG-3′ for SoyFERH1, and forward primer 5′-ACGTCGACTCGTTTTTCTTCCCAAATGG-3′ and reverse primer 5′-ACCTGCAGGGCCGTTCAAAGATTATACA-3′ for SoyFERH2. The OsFER1C cDNA sequence was amplified using forward primer 5′-TGCTGCAGCCTTTCCGCCATGCTTCCT-3′ and reverse primer 5′-TGACTAGTCCCATGGATGGAAGAAACGA-3′ while the OsFER2C cDNA sequence was amplified using forward primer 5′-TGCTGCAGATGCTTCCTCCTAGGGTTGC -3′ and reverse primer 5′-TGACTAGTCCATGGATGGAAGAAACGAA-3′. The ferritin cDNAs were then cloned separately in pCR 2.1 TOPO (Invitrogen), namely as pTOPO–SoyFERH1, pTOPO–SoyFERH2, pTOPO–OsFER1C, and pTOPO–OsFER2C. The cDNA clones were sent for sequencing (Macrogen, Korea) using universal primers M13 forward and M13 reverse. The sequences of the ferritin cDNA fragment were verified by alignment with known sequences of SoyFERH1 (NCBI accession no. M64337), SoyFERH2 (NCBI accession no. AB062754.1), OsFER1C (NCBI accession no. AF519570.1), and OsFER2C (NCBI accession no. AF519571.1).
Promoter cassettes were cut from pTOPO using restriction enzymes of EcoRI and BamHI and cloned into the corresponding sites of pCAMBIA1380 with hygromycin phosphotransferase (HPT) selectable marker for one-vector transformation and pCAMBIA0380 without HPT selectable marker for two-vector co-transformation. Ferritin cDNAs that were first cloned into pTOPO vectors were then isolated using the restriction sites (SalI and PstI for SoyFERH1 and SoyFERH2; PstI and SpeI for OsFER1C and OsFER2C, respectively) and inserted into the corresponding enzyme sites downstream of the promoters.
Production of transgenic plants with ferritin genes
The vectors were introduced into mega-variety IR64 by Agrobacterium tumefaciens strain LBA 4404. Rice immature embryos were co-cultivated with Agrobacterium following the modified procedure of Hiei and Komari (2006). For single transformation, the Agrobacterium strain used harbors a single transformation vector containing both the HPT and ferritin genes. For co-transformation to generate marker-free plants, a 5:1 mix ratio of Agrobacterium cultures of pCAMBIA 0380 containing the ferritin gene and pCAMBIA 1300 containing the HPT cassette was used.
Molecular evaluation for the presence of transgenes
Transgenic rice lines were characterized for initial screening by PCR by using a primer set of the ferritin gene and HPT gene (forward primer 5′-TACTTCTACACAGCCATC-3′ and reverse primer 5′-TATGTCCTGCGGGTAAAT-3′). Positive plants were analyzed for the presence of a single-copy insert by DNA blotting. Rice genomic DNA was extracted from leaves using the procedure described by Dellaporta et al. (1983). Hybridization probes were chemically labeled with digoxigenin using a PCR DIG probe synthesis kit (Roche Diagnostic GmBH, Mannheim, Germany). Prehybridization, hybridization, and detection were carried out following the manufacturer’s instructions (Roche Diagnostic GmBH).
Plants with a single copy of the gene of interest and fertile plants were selected for biochemical and phenotypic evaluation.
Phenotypic evaluation of transgenic plants
Phenotypes of the T1 and T2 single-copy events and their respective nulls were evaluated in the biosafety screenhouse. The marker-free plants were selected based on the presence of the ferritin gene and absence of HPT. T1 plants were transplanted following a randomized block design with three replicates. Each replicate of single events consists of seven PCR-positive and three null segregants and the wild type. Six parameters were recorded: (1) days to flowering, (2) plant height, (3) tiller number, (4) panicle number, (5) panicle length, and (6) panicle fertility. Days to flowering were counted from the start of sowing, while the length of the plant starting from above the soil to the tip of the tallest panicle was measured in centimeters for panicle length. For panicle fertility, all the panicles per plant were threshed; the filled and unfilled grains were separated using a grain blower and were counted afterwards. The data were analyzed statistically using Statistical Analysis Software (SAS) to determine whether there was a significant difference between the transgenic, null, and wild-type plants.
Grain quality evaluation of high-Fe seeds
Seed samples of transgenic and non-transgenic plants were evaluated for the traits of amylose content, cooking quality, protein content, and milling potential score in IRRI’s Grain Quality and Nutrition Center Laboratory.
Seed polishing method
Thirty brown rice grains were placed in 2-ml labeled micro tubes. The tubes were placed on a cryogenic metal rack (48 tubes per rack). The Genogrinder-2000 (SPEX CertiPrep Inc., Metuchen, NJ, USA) was set at 1,200 strokes min−1 for 2 min’ duration for 50 times. The samples were then placed in a new tube and run for an additional 20 times. Any trace of loose aleurone was removed.
Homozygous T2 screening
Forty-five events were selected based on phenotypic performance and Fe concentration for T2 homozygous screening. Homozygous lines were determined based on a 90–100 % resistance response of 30 randomly selected seeds of individual lines in 40 mg/L hygromycin solution.
In addition, 20 plants of each T2 line were checked by PCR using the primers of target genes. The DNA sequences of the PCR primers used in this study are the following:
-
SoyFERH1F: 5′-ATGGCTCTTGCTCCATCCAAAGTT-3′
-
SoyFERH1R: 5′-TTGATCAAAGTGCCAAACACCGTG-3′
-
SoyFERH2F: 5′-ACGTCGACTCGTTTTTCTTCCCAAATGG-3′
-
SoyFERH2R: 5′-ACCTGCAGCGCCGTTCAAAGATTATACA-3′
Iron concentration determination by ICP method
Polished seeds were sent to the Analytical Service Lab at IRRI for inductively coupled plasma–optical emission spectrometer (ICP-OES) analysis, and selected samples were validated by sending them to the Waite Analytical Laboratory, School of Agriculture, Food and Wine, University of Adelaide, Australia, for micronutrient concentration. Ten seeds (about 200 mg) from each plant were randomly selected, ground and wet-ashed with 2 ml HNO3 and H2O2 overnight at 110 °C. Ashing was repeated until the samples whitened. These samples were then dissolved in 15 ml of 1 N HCl. Concentration of Fe was measured using ICP-OES (Perkin Elmer ICP Optima 5300DV, Perkin Elmer, MA, USA) at 238.204 nm (Fe).
ELISA, in situ Western, and Western blotting for ferritin protein expression
Total protein was extracted from transgenic plants and non-transgenic control plants. For ELISA, 96-well microtiter plates (Linbro, VA, USA) were coated with 100 μl dilution of purified crude protein (25 μg) from transgenic and control plants in sodium bicarbonate buffer (pH 8.6), and were incubated at 4 °C overnight. After blocking with 100 μl of blocking solution (10 mg ml−1 bovine serum albumin in Tris-buffered saline with 0.05 % Tween 20 (TBST) for 120 min at room temperature, the plates were washed six times with TBST. One hundred μl of primary antibody, rabbit anti-SoyFERH1 serum (1:4,000), was added and the plates gently shaken for 1 h. Plates were again washed as described above and 100 μl TBST with diluted (1:2,000) goat anti-rabbit IgG conjugated with horseradish peroxidase (Bio-Rad, Hercules, CA, USA) was added as a secondary antibody and the plates were incubated for 60 min. The plates were rewashed as described above, and 200 μl well−1 of 3,3′,5,5′-tetramethylbenzidine (TMB) was added. After 30 min incubation at room temperature, the reaction was stopped by adding 100 μl of 3 M HCl. The optical density (OD) of each reaction was measured at 450 nm.
For in situ Western hybridization, mature polished seeds were soaked in distilled water at 4 °C overnight. The softened seeds were sectioned longitudinally with a razor blade. The Qu et al. (2005) protocol was followed for SoyFERH1 distribution in rice seeds.
Western blot analysis was carried out as described previously (Datta et al., 1997) using rabbit anti-SoyFERH1 serum.
Iron staining for localization of Fe
Perls’ Prussian blue technique was employed for the localization of Fe. Polished whole rice seeds were soaked in water in a 1.5-ml microfuge tube overnight. Seeds were sectioned in a Petri dish using a ceramic knife. Whole and sectioned seeds were stained with 2 % HCl (Merck, Germany) and 2 % potassium ferrocyanide (Sigma, MO, USA) to form an insoluble blue color after reaction with Fe. The seeds were washed six times with water to remove excess stain. Observation and documentation were done with an Olympus SZX-7 stereomicroscope with digital camera DP 71 under bright-field mode.
Gene expression using real-time PCR
Gene expression analysis of the ferritin gene (SoyFERH1) was conducted at the milky and mature seed stages of transgenic indica mega-variety IR64. RNA extraction from full grain, primer design, and conditions were optimized for quantitative PCR (qPCR). The relative expression of ferritin genes was normalized using the housekeeping genes actin, ATPase, and GAPDH. Measurements were made in four biological and four technical replicates.
Gene expression analysis was employed to study the changes in expression due to the insertion of the soybean ferritin gene on the endogenous ferritin gene along with Fe transporter genes in seeds of transgenic indica cv IR64. Quantitative PCR was performed using primers designed for FRO1, FRO2, YSL18, SOYFERH1, NAAT, OsFer, YSL15, NAS1, NAS2, and NAS3 (Supplementary Table 1). A real-time PCR was prepared using a master mix of the following reaction components: 6 μl molecular biology grade PCR water, 1 μl forward primer (0.25 μM), 1 μl reverse primer (0.25 μM), 10.0 μl LightCycler® 480 SYBR Green I Master (Roche Diagnostics, Roche Applied Science), and 2 μl cDNA (50 ng reverse-transcribed DNA from total RNA, Roche Diagnostics) were added as a PCR template.
Results
Promoter selection
Three different endosperm-specific promoters, GLUB1, GLUB4, and GLB1, were evaluated for their specific expression in mega-variety IR64. The promoters were fused to the GUSA reporter gene to study their expression pattern. The aim was to obtain promoters that are primarily active during early embryo development to allow translocation of Fe.
GUS expression in the seed was observed only in the endosperm in all three promoters evaluated (Supplementary Fig. 1). Based on blue staining of several seeds, it was observed that, at the mature stage, expression was comparable in all three promoter constructs, but, for GLUB1-GUS seeds, a higher intensity of blue color was observed in the outer part of the endosperm, while GLB1 had a more even distribution of blue color in the endosperm, similar to the observation of Qu and Takaiwa (2004). At the early developmental dough stage, GLUB1 showed the highest expression of the GUS gene, followed by GLUB4 and GLB1. We selected the first two promoters for further study, based on the hypothesis that highly active promoter during early stage of grain formation will increase the chances of metal loading in the grain.
Production of transgenic plants with ferritin genes
A large number of independent transgenic events were produced from the integration of different combinations of two rice glutelin promoters with soybean and rice ferritin genes into IR64 with transformation frequency ranging from 10.8 to 38.1 % for single transformation and from 3.3 to 25.9 % for co-transformation (Table 1a, b). A total of 693 and 361 transgenic events from single and co-transformation experiments, respectively, were produced, and integration of the different ferritins was confirmed by PCR analysis with the specific corresponding primers. The percentage of single-copy insertions, analyzed by DNA blotting, was 31–87 % of the PCR-positive plants (Supplementary Fig. 2). Plants were grown to maturity and progenies of independent fertile single-copy transgenic lines of IR64 developed from different constructs were selected and grown in the screenhouse for mass screening to evaluate their Fe concentration.
PCR analysis in the T1 generation of selected co-transformed plants for the presence of the ferritin and absence of HPT genes showed that the marker-free genotype comprised about 7–25 % of the segregating progenies among the lines analyzed (Table 1b). Plants having both genes comprised about 50–90 % of the segregating progenies.
Phenotypic evaluation of transgenic plants
A total of 1,860 plants from 58 T1 IR64 single transformation transgenic events and 768 progenies from 27 co-transformation events with GLUB1:SoyFERH1, GLUB4:SoyFERH2, GLUB1:SoyFERH2, GLUB1:OsFER1C, GLUB1:OsFER2C, or GLUB4:OsFER2C were transplanted in the paddy soil screenhouse (Supplementary Fig. 3), consisting of seven T1-positive plants/events and three corresponding nulls (azygous) in three replicates. Agronomic characteristics, including fertility, plant height, seed weight, panicle number, and panicle length of the plants, were measured and evaluated at three different growth stages. Variation between mean measurements of the five parameters was analyzed statistically using SAS. The mean values of seed weight per plant and plant height among transgenics (single transformation) with different ferritin genes, their null segregants, and the wild-type IR64 were not significantly different at the 5 % level of significance (Table 2). The mean values of tiller number, panicle length, and panicle number increased in transgenics, and in their nulls. For co-transformation transgenic plants, the mean values of seed weight, plant height, and panicle length were not significantly different while the mean values of the tiller number and panicle number increased in the transgenics and their nulls (Table 2).
Grain quality evaluation of high-iron seeds
The grain quality of three events each from single transformation and co-transformation transgenic plants representing four different phytoferritin genes was found to be similar in milling and cooking quality to that of mega-variety IR64 in our grain quality evaluation study (Supplementary Table 2). The amylose content of the transgenic events analyzed ranged from low to intermediate. The gelatinization temperature of the transgenic seeds was scored as intermediate to high-intermediate. The transgenic and wild-type seeds had soft gel consistency, ranging from 85 to 100 mm in length. The protein content of the transgenic seeds was 10.1–12.6 %. In terms of milling characteristics, the milling potential score of the transgenic seeds was 1 and chalkiness ranged from 1 to 2, which is similar to that of the IR64 wild type.
Effect of the promoter and transgene on Fe concentration in the endosperm
Seeds from individual plants of 58 events of the T1 generation were polished by a Kett mill and analyzed for Fe concentration using a low-cost colorimetric assay, followed by ICP-OES at the IRRI Analytical Service Laboratory (ASL) for representative samples. Milled rice from 16 individual plants from each independent event was pre-screened for Fe content using a colorimetric assay. The Fe concentration of milled rice of selected events from each construct was then analyzed by ICP-OES. Supplementary Table 3 shows the comparison of the Fe concentration of IR64 plants expressing ferritin genes in the T2 generation driven by GLUB1 (5.63 ± 0.67 mg kg−1) and plants with ferritin genes regulated by GLUB4 (6.21 ± 0.74 mg kg−1). Fe concentration by colorimetric assay for transgenic plants with the OSFER2C gene was of a level similar to that of the wild-type IR64 (data not shown); these lines were therefore not advanced further for further analysis and evaluation.
Forty-five events were selected for the T2 homozygous screening. Homozygous lines were selected based on segregation analysis. We changed the polishing method to the Genogrinder (Bautista et al. 2004) in the succeeding generations to be more stringent in milling quality for a low number of seeds. The Fe concentration of wild-type IR64 using this instrument was consistently 2–3 mg kg−1, which is similar to observations by Batista et al. (2012) that Fe concentration of IR64 in several commercial mills ranged from 2 to 3 mg kg−1. Both GLUB1 and GLUB4 promoters showed a comparable increase in Fe in the transgenic seeds (Supplementary Table 3). In addition, ICP results of the T2 and T3 generation of transgenic seeds differed between different types of ferritin genes (OsFER1C, SoyFERH1, and SoyFERH2). However, the Fe concentration of transgenic seeds from OsFER1C- and SoyFERH2-transformed plants was significantly lower in the T3 ICP results than in plants transformed with SoyFERH1 (Supplementary Fig. 4).
Fe concentration in the polished transgenic seeds of the T3 generation of single and co-transformation events representing different constructs was measured using ICP at the Waite Analytical Laboratory in Adelaide (Fig. 1). The maximum value of Fe concentration in transgenic lines expressing the ferritin gene in homozygous lines was 7.6 mg kg−1 for GLUB4::SoyFERH1 (single transformation), followed by 5.9 mg kg−1 for GLUB4::SoyFERH1 (co-transformation). The transgenic T3 lines showed as much as a 3.4-fold increase in Fe concentration compared with the non-transgenic seeds.
Correlation of Fe concentration and ferritin expression
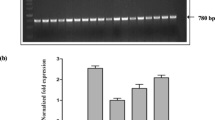
SoyFERH1 protein expression in T2 and T3 seeds of a number of selected events driven by GLUB1 and GLUB4 promoters was studied by ELISA, using rabbit anti-SoyFERH1 antibody. The ELISA fold values were calculated using IR64 wild type as background value and were compared with the Fe ICP values. Transgenic events with higher ferritin expression in ELISA showed higher Fe using ICP in both the T2 (data not shown) and T3 generations (Fig. 2a).
Expression of ferritin genes. a Comparison of fold increase in SoyFERH1 protein and Fe concentration in transgenic plants in T3 generation ELISA (blue bars represent mean fold change ± standard error obtained from three technical replicates). Data are illustrated as fold change in transgenic seeds relative to baseline values (wild-type IR64) Fold increases in Fe content (magenta bars represent mean fold change ± standard error) in seeds of transgenic plants over that of wild type were measured from three biological replicates. b Western blot analysis of SoyFERH1 ferritin in transgenic rice seeds. Fifty micrograms of total proteins extracted from each transgenic and non-transgenic seed were fractioned by SDS-PAGE, immunoblotted, and then bound with soybean ferritin rabbit polyclonal antibodies. The predicted 28-kDa protein band is ferritin. M = Protein Size Marker (Amersham, RPN 756); WT = wild-type IR64; 1–4, 6–7 = T1 IR64 events with SoyFERH1; 5 and 8 are null segregants. c In situ Western blot of transgenic rice seeds with ferritin. (I–II) Polished seed (longitudinal section and whole seed) of IR64 transformed with SoyFERH1 driven by GLUB4 promoter bound with anti-SoyFERH1 antibody (left) versus polished IR64 seed (right). (III–IV) Polished seed (longitudinal section and whole seed) of marker-free IR64 transformed with SoyFERH1 driven by GLUB1 promoter bound with anti-SoyFERH1antibody (left) versus polished IR64 seed (right). (V–VI) Fe localization using Pearl Prussian blue staining (transverse section and whole polished seed of transgenic IR64 with SoyFERH1 driven by GLUB4 (left) versus wild-type IR64 (right). (VII–VIII) Fe localization using Pearl Prussian blue (transverse section and whole polished seed of transgenic marker-free IR64 with SoyFERH1 driven by GLUB1 (left) versus wild-type IR64 (right)
Ferritin localization and iron distribution in transformed lines
The expression pattern of soybean ferritin directed by a rice endosperm-specific promoter (GLUB1 or GLUB4) was determined by in situ Western hybridization. In IR64 plants with SoyFERH1 driven by GLUB4, color development was more homogeneous in the endosperm with higher concentrations in its outer cells (Fig. 2c I–IV). In IR64 transgenic T2 and T3 plants with SoyFERH1 directed by the GLUB1 promoter, ferritin was expressed in a localized part of the starchy endosperm tissue. Non-transformed (NT) rice seed controls remained minimally stained compared with the transgenic lines.
Prussian blue staining clearly shows the accumulation of Fe in the endosperm cells of polished transgenic rice grains (T2 and T3), as indicated by the blue color (Fig. 2c V–VIII). In non-transgenic rice grains, the endosperm showed minimal color development.
Western blot analysis of total protein extracts from mature polished seed from T1 plants, null segregants, and the wild type was performed to investigate the expression of ferritin (Fig. 2b). Polyclonal antibody directed against SoyFERH1 ferritin was bound to a 28-kDa band from transgenic plants transformed with the SoyFERH1 gene, while no 28-kDa band was observed in proteins from the NT and the null segregants from the T1 trangenic lines. Though ferritin amounts varied between different individual lines, the protein could be observed in almost all the transformants assayed. In addition, another band, about 22 kDa, was observed in transgenic rice, although at a lower intensity in the wild-type and null segregants of the transgenic lines, which could be the native rice ferritin (23–24 kDa).
Gene expression study of ferritin genes
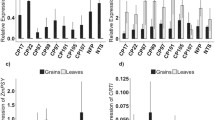
To examine the expression of the ferritin gene, total RNA isolated from T2 immature seeds of six lines (single and co-transformation events) was analyzed by qPCR using primers specific for soybean ferritin. The increase in expression of the SoyFERH1 gene of the different events ranged from 3- to 37-fold (Fig. 3a).
The expression of Fe transporter genes FRO1, FRO2, YSL16, YSL18, NAS1, NAS2, and NAS3 and ferritin genes SoyFERH1 and SoyFERH2 was analyzed in selected IR64 transgenic plants and compared with the expression of those genes in IR64 seeds in the mature stage. Expression of the endogenous transporter genes involved in cellular delivery of Fe generally increased in the three transgenic lines, particularly for NAS2, NAS3, YSL15, and YSL18 genes (Fig. 3b) and downregulation of Fe was observed in the transgenic events.
Discussion
The high transformation efficiency in both single transformation and co-transformation of mega-variety IR64 allowed the evaluation of multiple lines for agronomic traits and eventual selection of lines with higher iron concentration. Phenotypic data of plants from independent transgenic events with a single insertion of the ferritin gene in paddy soil conditions showed similar seed weight per plant, plant height (for single and co-transformation), and panicle length (co-transformation) among transgenic, null, and wild-type plants, indicating no effect of the genes on these traits. However, variations were observed in tiller number, panicle number, and panicle length (for single transformants) in both transgenic and null segregrants, most likely due to an effect of environment or tissue culture rather than a gene effect. We also did not see any chlorotic symptom in the leaves as reported by Qu et al. (2005).
Grain quality evaluation of seeds from transgenic plants shows that seeds with high Fe are comparable in milling characteristics and cooking traits with non-genetically modified IR64 seeds. IR64 possesses good market value because of its preferred milling properties and good cooking quality; transgenic rice with ferritin genes could thus offer additional nutritional value to this preferred variety. Our results for milling potential and chalkiness with advanced transgenic lines with SoyFERH1, SoyFERH2, and OsFER1C indicate that the transgenic seeds have high milling quality from a marketing standpoint. Based on the adoption rate of IR64 with submergence tolerance in Southeast Asia (Manzanilla et al. 2011), biofortification by the introduction of a transgene into mega-variety IR64 will increase the chances of quicker adoption.
GLUB1 and GLUB4 promoters were selected for expressing plant ferritins in transformation to allow translocation of Fe from leaves to the endosperm in an early seed development stage. However, we found that, even though ferritin gene expression could be increased up to 37-fold using GLUB1 and GLUB4, similar to the GLB1 promoter study of Qu et al. (2005), the maximum Fe accumulation was only threefold that of the wild type. Increasing sink at early grain development stage could therefore only moderately increase mature grain Fe. Gene expression analysis showed that the introduction of the SoyFER gene in rice increases gene expression of metal transporters yellow stripe-like (YSL15 and YSL18) and nicotinamine synthase (NAS2 and NAS3). Nicotianamine (NA) is thought to be an essential chelator for metal homeostasis and plays key roles in Fe metabolism and homeostasis in all higher plants. Nicotianamine synthase catalyzes the trimerization of S-adenosylmethionine to form one molecule of NA. Apparently, an increase in Fe storage protein gives a signal to other Fe homeostasis genes to increase the rate of Fe transport to the seed; in other words increasing the sink upregulates the genes involves in mobilizing the source, but a decrease in native ferritin levels was observed. However, despite the high availability of storage protein in the grain and increased expression of the Fe homeostasis gene in the grain, limited additional Fe was loaded to the grain, showing the need for stronger mobilization of Fe from vegetative tissue to the grain.
Free Fe in cells is toxic and strict control of Fe homeostasis is required to avoid deficiency and toxicity. Ferritin plays a role in both Fe housekeeping and storage and also in Fe detoxification. Moreover, the physiological role of endogenous ferritin appears to be more related to protection against excess Fe than to reserve storage (Ravet et al. 2009a).
We report the generation of marker-free transgenic rice carrying ferritin genes driven by endosperm promoters, and its evaluation for increased Fe in the seed and agronomic analysis. Stable inheritance was confirmed by progeny analysis of the ferritin genes according to the Mendelian (3:1) ratio. PCR analyses of T1 plants show 7–25 % marker-free plants with high Fe (5.9 mg kg−1) in the seeds. This indicates that co-transformation using two Agrobacterium strain individually harboring the HPT or ferritin genes resulted in integration in different loci of the two genes in most co-transformation events. This separation allowed the segregation of the genes in the next generation as planned. Genetically modified plants without antibiotic-selectable markers are likely to be more acceptable by the public, and also with lower numbers of transgenes there would be a reduced requirement for biosafety evaluation of the novel protein in the transgenic plant. In the genetic background of IR64, a popular indica variety with Fe concentration 2–3 mg kg−1, we obtained a concentration based on ICP as high as 7.2 ± 1.52 mg kg−1 in single-copy T2-generation polished seed (data not shown), slightly higher than the Fe concentration previously reported by Wirth et al. (2009), with 7 mg kg−1 in events with single-copy insertion. This accounts for a 2.5-fold increase in extra Fe compared to the control. The highest Fe concentration of a T3 transgenic IR64 event expressing the ferritin gene (7.6 mg kg−1) was obtained from GLUB4::SoyFERH1 (single transformation-homozygous line). A transgenic line with SoyFERH2 driven by GLUB1 had an Fe concentration of 2.9 mg kg−1. SoyFERH1- and SoyFERH2-transformed lines had much higher Fe concentration than the line with the OsFER1C gene. In co-transformation lines, the highest Fe concentration obtained was 5.9 mg kg−1 from one of the GLUB4::SoyFERH1 plants. Sequence comparison of two soybean and two rice ferritins showed high similarity in the α, β, and D helical regions but they differed in the C helical region (Supplementary Fig. 5). In addition to this difference, the transit peptide region of the sequences showed only low sequence similarity. This may also cause a difference in their ability to accumulate Fe in the grain. Introduction of an extra copy of endogenous rice ferritin can also trigger post-transcriptional gene silencing (Vaucheret et al. 2001) due to the specific degradation of a population of homologous RNAs, and result in a minimal increase in or even reduction of the ferritin content. The extra copy of endogenous ferritin may also cause tighter control of Fe homeostasis compared with exogenous ferritin, since one major role of ferritin is to reduce Fe-mediated oxidative stress, apart from functioning as a Fe storage protein (Ravet et al. 2009a, b).
The efficacy of GLUB1 and GLUB4 endosperm-specific promoters, both more active in the early stage of grain filling, in increasing ferritin expression and subsequently Fe concentration in polished grain is not consistently different among different generations of different constructs, based on ICP and spectrophotometric quantification and ELISA using SoyFERH1 antibody.
The high-Fe lines consistently showed high protein expression using ELISA. Transgenic lines with a six to sevenfold increase in ferritin protein expression determined by ELISA exhibited an average threefold increase in Fe concentration as determined by ICP-OES. Aluru et al. (2011) also reported a strong correlation between ferritin concentration and Fe content in transgenic maize with soybean ferritin. Qu et al. (2005) observed that the fold increase in the amount of ferritin storage estimated using Western blotting was not linear with the increase in Fe concentration. From the population analyzed in this study, the lines expressing higher ferritin had higher Fe.
Iron translocation from leaf tissue and other plant tissues to seed endosperm is known to be one of the major limiting factors in a biofortification approach. Iron is incorporated into the ferritin shell to form the mineral core in the plastids (Waldo et al. 1995). Although the concentration of Fe measured in elite transgenic IR64 (7 mg kg−1) was higher than with the two-gene approach (Wirth et al. 2009), it is clear that increasing the sink should be coupled with an improvement in Fe loading to the amyloplast, where the ferritin is stored in the seed. Goto et al. (1999) stated that proper assembly of the protein is required for import of the subunit into the plastid, as well as for Fe storage function.
Another issue regarding over-expressing ferritin for accumulating Fe in the grain is the function of H1 and H2 subunits, according to Deng et al. (2010). Heteropolymeric ferritin may facilitate plant cell absorption of both ferrous and ferric ions from soil more effectively than homopolymeric ferritin. Both Fu et al. (2010) and Deng et al. (2010) reported that the H2 subunit is more resistant to proteolysis than the H1 subunit, but we saw only a slight increase in Fe with events expressing H1 instead of H2. If H1 and H2 act synergistically, expressing both could result in better Fe absorption.
Preliminary results also show that the introduction of the soybean ferritin gene reduces the expression of endogenous genes. Transgenic events expressing NAS2 (Johnson et al. 2011) and the activation of NAS2 and NAS3 genes (Lee et al. 2009, 2012) have been shown to increase Fe content in rice. These studies strongly support the importance of both Fe homeostasis genes in Fe grain filling. In our study, over-expression of exogenous ferritin by itself did not overcome the limiting rate of Fe uptake to the endosperm; however, NAS2 is a potential gene for pyramiding with other genes to improve Fe concentration in endosperm since it does not cause any detrimental effect on plant performance. Pyramiding genes have been reported by Masuda et al. (2012) and Wirth et al. (2009), where OsNAS1 was used in both cases. Our results shows that strong upregulation was detected in OsNAS2, which was confirmed by previous studies (Lee et al. 2009 and Johnson et al. 2011); for the multigene gene approach OsNAS2 and OsNAS3 may thus have more potential than OsNAS1.
In summary, our study demonstrated that the introduction and expression of ferritin genes from soybean and rice under the control of endosperm-specific promoters increased the concentration of iron in polished seeds in an important rice variety, IR64, and in its progenies without compromising the agronomic and grain quality of the transgenic plants. Expressing ferritin from soybean was more effective in increasing the iron content in transgenic rice than over-expression of ferritin from rice. Endosperm-specific promoter GLUB4 resulted in a higher iron concentration in transgenic plants than GLUB1, regardless of phytoferritin genes being expressed. Introduction of the ferritin gene is still a potential approach to develop a transgenic product in combination with other gene/s that will allow additional iron loading in the grain to meet the target of fulfilling a significant part of the estimated average requirement of iron in the human diet.
References
Aluru M, Rodermel S, Reddy M (2011) Genetic modification of low phytic acid 1–1 maize to enhance iron content iron content and bioavaibility. J Agric Food Chem 59:12954–12962
Bajaj S, Mohanty A (2005) Recent advances in rice biotechnology-towards genetically superior transgenic rice. Plant Biotechnol J 3:275–307
Batista B, Nacano L, De Freitas R, De Oliveira-Souza V, Barbosa F (2012) Determination of essential (Ca, Fe, I, K, Mo) and toxic elements (Hg, Pb) in Brazilian rice grains and estimation of reference daily intake. Food and Nutrition Sciences 3:129–134
Bautista B, Siebenmorgen TJ, Mauromustakos A, Burgos RM (2004) Small sample mill protocol development: evaluation of a Genogrinder 2000. In: Norman RJ, Meullenet JF, Moldenhauer KAK (eds) B.R. wells rice research studies 2004, Univ. of Ark., Agric. Exp. Stn. Res. Ser. 529. Arkansas, pp 357–363
Beard JL, Burton JW, Theil EC (1996) Purified ferritin and soybean meal can be sources of iron for treating iron deficiency in rats. J Nutr 126:154–160
Briat JF, Lobréaux S (1997) Iron transport and storage in plants. Trends Plant Sci 2:187–193
Briat JF, Duc C, Ravet K, Gaymard F (2010) Ferritins and iron storage in plants. Biochim Biophys Acta 1800:806–814
CERA (2010) GM Crop Database. Center for environmental risk assessment (CERA), ILSI research foundation, Washington DC, USA. http://cera-gmc.org/index.php?action=gm_crop_database. Accessed 13 Sep 2012
Copenhagen Consensus (2004) Addresses 10 major challenges in the world. http://www.copenhagenconsensus.com. Copenhagen Consensus Center—CCC Home Page. Accessed 12 Oct 2012
Datta SK, Torrizo LB, Tu J, Oliva NP, Datta K (1997) Production and molecular evaluation of transgenic rice plants. In: IRRI discussion paper series no. 21. International Rice Research Institute, Manila, pp 35–39
Davila-Hicks P, Theil EC, Lonnerdal B (2004) Iron in ferritin or in salts (ferrous sulfate) is equally bioavailable in nonanemic women. Am J Clin Nutr 80:936–940
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Deng J, Cheng J, Liao X, Zhang T, Leng X, Zhao G (2010) Comparative study on iron release from soybean (Glycine max) seed ferritin induced by anthocyanins and ascorbate. J Agric Food Chem 58:635–641
Drakaki G, Marcel S, Glahn RP, Lund EK, Paraigh S, Fischer R, Christou P, Stoger E (2005) Endosperm-specific co-expression of recombinant soybean ferritin and Aspergillus phytase in maize results in significant increases in the levels of bioavailable iron. Plant Mol Biol 59:869–880
European Food Safety Authority Scientific Panel (2004) Opinion of the scientific panel on genetically modified organisms on the use of antibiotic resistance genes as marker genes in genetically modified plants. EFSA J 48:1–18
Fu X, Jianjun D, Haixia Y, Masuda T, Goto F, Yoshihara T, Zhao G (2010) A novel EP-involved pathway for iron release from soya bean seed ferritin. Biochem J 427:313–321
Ghandilyan A, Vreugdenhil D, Aarts MGM (2006) Progress in the genetic understanding of plant iron and zinc nutrition. Physiol Plant 126:407–417
Goldstein DA, Tinland B, Gilbertson LA, Staub JM, Bannon GA, Goodman RE, McCoy RL, Silvanovich A (2005) Human safety and genetically modified plants: a review of antibiotic resistance markers and future transformation selection technologies. J Applied Microbiol 99:7–23
Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nature Biotechnol 17:282–286
Hare P, Chua NH (2002) Excision of selectable marker genes from plants. Nature Biotechnol 20:575–580
Hiei Y, Komari T (2006) Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tissue Org Culture 85:271–283
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: B-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Johnson AAT, Kyriacou B, Callahan DL, Carruthers L, Stangoulis J, Lombi E, Tester M (2011) Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS ONE. doi:10.1371/journal.pone.0024476
Khush GS (2005) What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol 59:1–6
Kondrák M, van der Meer IM, Bánfalvi Z (2006) Generation of marker- and backbone-free transgenic potatoes by site-specific recombination and a bi-functional marker gene in a non-regular one-border agrobacterium transformation vector. Transgenic Res 15:729–737
Lee S, Jeon US, Lee SJ, Kim YK, Persson DP, Husted S, Schjørring JK, Kakei Y, Masuda H, Nishizawa NK, An G (2009) Iron fortification of rice through activation of the nicotianamine synthase gene. Proc Natl Acad Sci USA 106:22014–22019
Lee S, Kim YS, Kim YK, Schjeriring JK, An G (2012) Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol Cells 33:269–275. doi:10.1007/s10059-012-2231-3
Lönnerdal B (2009) Soybean ferritin: implications for iron status of vegetarians. Am J Clin Nutr 89:1680S–1685S
Manzanilla DO, Paris TR, Vergara GV, Ismail AM, Pandey S, Labios RV, Tatlonghari GT, Acda RD, Chi TTN, Duoangsila K, Siliphouthone I, Manikmas MOA, Mackill DJ (2011) Submergence risks and farmers’ preferences: implications for breeding Sub1 rice in Southeast Asia. Agric Syst 104:335–347
Masuda H, Ishimaru Y, Aung MS, Kobayashi T, Kakei Y, Takahashi M, Higuchi K, Nakanishi H, Nishizawa NK (2012) Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci Rep. 2:543
Miki B, McHugh S (2004) Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J Biotechnol 107:193–232
Murray-Kolb LE, Takaiwa F, Goto F, Yoshihara T, Theil EC, Beard JL (2002) Transgenic rice is a source of iron for iron-depleted rats. J Nutr 132:957–960
Paul S, Ali N, Gayen D, Datta SK, Datta K (2012) Molecular breeding of Osfer2 gene to increase iron nutrition in rice grain. GM Crops Food 3:310–316
Qu Q, Takaiwa F (2004) Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J 2:113–125
Qu Q, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222:225–233
Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F (2009a) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57:400–412
Ravet K, Touraine B, Kim SA, Cellier F, Thomine S, Guerinot M, Briat JF, Gaymard F (2009b) Post-translational regulation of AtFER2 ferritin in response to intracellular iron trafficking during fruit development in Arabidopsis. Mol Plant 2:1095–1106
Theil EC (1987) Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem 56:289–315
Vasconcelos M, Datta K, Oliva N, Khalekuzzaman M, Torrizo L, Krishnan S, Olivera M, Goto F, Datta SK (2003) Enhanced iron and zinc accumulation in transgenic rice with the ferritin gene. Plant Sci 164:371–378
Vaucheret H, Beclin C, Fagard M (2001) Post-transcriptional gene silencing in plants. J Cell Sci 114:3083–3091
Waldo GS, Wright E, Whang ZH, Briat JF, Theil EC, Sayers DE (1995) Formation of the ferritin iron mineral occurs in plastids. Plant Physiol 109:797–802
Wirth J, Poletti S, Aeschlimann B, Yakandawala N, Drosse B, Osorio S, Tohge T, Fernie A, Gunther D, Gruissem W, Sautter C (2009) Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol J 7:631–644
World Bank (2006) Repositioning nutrition as central to development: a strategy for large scale action. World Bank, Washington, DC
World Health Organization (2001) Iron deficiency anaemia: assessment, prevention, and control: a guide for programme managers. World Health Organization (WHO/NHD/01.3), Geneva
World Health Organization (2008) Iron deficiency anaemia: assessment, prevention, and control: a guide for programme managers. In: de Benoist B, McLean E, Egli I, Cogswell M (eds) Worldwide prevalence of anaemia 1993–2005: WHO global database on anaemia. World Health Organization (WHO/NHD/01.3), Geneva
Zhao FJ, Shewry PR (2011) Recent developments in modifying crops and agronomic practice to improve human health. Food Policy 36(Supplement 1):S94–S101
Acknowledgments
This study was funded by HarvestPlus and by USAID. Fumiyuki Goto kindly provided the anti-SoyFERH1 polyclonal antibody. We would like to thank IRRI statistician, Violeta Bartolome, for helping in the statistical analysis and B. Hardy for editing this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Norman Oliva and Prabhjit Chadha-Mohanty contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Oliva, N., Chadha-Mohanty, P., Poletti, S. et al. Large-scale production and evaluation of marker-free indica rice IR64 expressing phytoferritin genes. Mol Breeding 33, 23–37 (2014). https://doi.org/10.1007/s11032-013-9931-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9931-z