Abstract
Seedling establishment is a vulnerable stage in the crop life cycle which can be affected by different abiotic stresses. Drought and salinity are major environmental constraints worldwide, but few studies have genetically compared these two stresses on the same genetic material. In this study, the dynamic response of barley seedling growth to different levels of salt stress and PEG6000-mediated osmotic stress was evaluated in a European winter barley cultivar collection, in parallel with non-stress conditions. Salt and osmotic stress experiments produced different root-response curves. A final set of 56 phenotypic traits was subjected to genome-wide association mapping with 4885 gene-based SNP markers: 28 quantitative trait loci (QTL) were identified; 10 loci were found to be involved in saline conditions, whereas 20 loci were detected under osmotic stress. Four loci on chromosomes 1H, 5H and 6H were detected under more than one growth condition. One co-localized QTL was involved in both root and shoot growth only under salt stress. A set of potential candidate genes with putative pleiotropic effects on seedling growth under different conditions is proposed, based on their physical proximity to the QTL peak markers. To conclude, we found that a QTL controlling seedling growth under one abiotic stress can respond to another stress. QTL promoting faster growth under non-stress conditions were also identified under salt and/or osmotic stresses. These loci might be exploited in breeding programs to overcome environmental stresses at the initial seedling stage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drought and salinity are major abiotic stresses limiting crop productivity (Rizwan et al. 2015). Climate change is expected to significantly impact temperature and precipitation profiles, thus increasing the incidence and severity of environmental stresses (Mondini and Pagnotta 2015). For this reason, drought stress is often combined with salinity in many of the world’s arid and semiarid regions, where irrigation water is prone to leach down through the salt patches on the soil surface (Sun et al. 2015).
Agricultural drought is defined as a period with decreased soil moisture that leads to crop failure without any reference to surface water resources (Mishra and Singh 2010). It is quantified as a reduction in water potential (ψw) and classified as an osmotic threat (Mondini and Pagnotta 2015). This subsequently limits nutrient uptake and photosynthetic assimilation by crops to achieve better productivity (Shi et al. 2015). In addition to osmotic stress, plants exposed to high salinity also experience ion toxicity (Zhang et al. 2009). Under high-saline conditions, osmotic adjustments are suggested to be triggered first (Zhu et al. 2015), then the plant regulates cellular ion homeostasis through the actions of multiple transporters in response to ion toxicity (Deinlein et al. 2014), while osmolyte adjustment is a key element in drought tolerance (Cattivelli et al. 2008). Given the common osmotic aspect, it is not surprising that a number of stress-regulated transcription factors are activated in response to both salt and drought stress, such as MYB (Dai et al. 2007), bZIP (Ying et al. 2012), AP2 (Zhang et al. 2009), and WRKY (Tao et al. 2011). Nevertheless, most reports have focused on testing these genes’ functions rather than on their exploitation in breeding programs for stress tolerance (Yun et al. 2012).
Genetic analysis of the response to salt and drought stress in rice identified five quantitative trait loci (QTL) controlling tolerance to both conditions, with two of them explaining a large portion of the observed phenotypic variation (Yun et al. 2012). Similarly, a major QTL for terminal drought tolerance in pearl millet was also associated with reduced salt uptake and enhanced growth under salt stress (Sharma et al. 2011). Further studies on the same genetic materials showed that drought-tolerant pearl millet genotypes maintain higher Na+ content in roots and old leaves, and higher chlorophyll and proline concentrations in younger leaves, relative to drought-sensitive lines (Sharma et al. 2014). The major adjustments in ion distribution and osmolytes in drought-tolerant lines were also associated with higher yields.
Seed germination is a developmental process triggered by embryo imbibition and resulting in radicle and coleoptile emergence. Germination supports the subsequent seedling growth, thus playing an important role in plant development and final yield in crops (Honsdorf et al. 2014). Unfavorable soil conditions, such as drought and high salinity, can significantly reduce seed-germination rate (Chloupek et al. 2003; Zhu et al. 2017). Therefore, specific crop management approaches have been suggested to improve seedling emergence in arid and saline soils, such as deep-sowing in dry lands (Abdel-Ghani et al. 2015; Takahashi et al. 2008) and seed priming for saline–alkali soils (Rashid et al. 2006). Under experimental conditions aimed at studying seed germination or seedling growth, saline conditions and the osmotic component of drought stress are frequently simulated by sodium chloride (NaCl) and polyethylene glycol (PEG), respectively (Abdel-Ghani et al. 2015; Shi et al. 2015). To date, two major QTL controlling the response of barley seedlings to salt stress have been identified on chromosome 5HL in the Oregon Wolfe Barley (OWB) mapping population (Witzel et al. 2010), and on chromosome 7HS in a Nure × Tremois (NT) population (Xue et al. 2017). In contrast, only minor QTL were detected when seedlings of the OWB population were exposed to osmotic stress (Bálint et al. 2008), and similar results were observed for other recombinant populations (Wójcik-Jagła et al. 2013). Genome-wide association (GWA) studies were also run in barley to study the genetic bases of tolerance to salt (Fan et al. 2016; Long et al. 2013; Sbei et al. 2014) and drought stress (Varshney et al. 2012; Wehner et al. 2015), mainly focusing on the vegetative stage rather than the seedling stage.
Different experimental methodologies have been adopted to phenotype barley seedlings under salt and PEG-mediated osmotic stress conditions, such as exposure to 150/250 mM NaCl (Witzel et al. 2010) or to 15/18% PEG (Abdel-Ghani et al. 2015). Tolerance indices/scores are often calculated as percentage of plant growth under stress conditions compared with controls (Abdel-Ghani et al. 2015; Tavakkoli et al. 2012). The mathematical Sigmoid model has also been applied to depict the dynamic response of seminal root growth in maize seedlings exposed to different salinities (Khan et al. 2003). Similarly, the Hill function of the Sigmoid model was employed to simulate the response of barley seedlings to salt stress, and key parameters from the simulation curves (e.g., maximum seedling length (Lmax), salt concentrations leading to 50% reduction in Lmax (Salt50)) were extracted for QTL mapping (Xue et al. 2017). To date, these methodologies have been used separately for genetic analyses; however, their combination might increase the accuracy and efficiency of QTL discovery.
A diverse barley collection of European winter cultivars was previously exploited to study the genetic bases of flowering time and leaf size (Digel et al. 2016), and part of the collection was also investigated for grain and straw yield (Bellucci et al. 2017). In the present study, the same population was phenotyped with a vertical germination system to construct dynamic simulation curves of seedling growth in response to different NaCl and PEG6000 treatments. The key parameters of salt-induced and PEG-mediated seedling response curves were employed for GWA mapping, in parallel with phenotypic data from individual stress treatments and control conditions. QTL controlling seedling responses to salt and osmotic stress were then compared with those controlling seedling growth in the absence of stress to separate the general effect on seedling growth from the specific effects controlling growth under stress conditions. The mapping procedure has been illustrated in Supplementary Figure 1. Four QTL were highlighted, one with a specific effect on stress-induced responses and the other three with a general effect on seedling development. In addition, two QTL regions, on chromosomes 1H and 5H, were further characterized for the presence of potential candidate genes involved in the traits under study.
Materials and methods
Plant material and phenotyping
A barley collection composed of 143 European winter cultivars, a part of the germplasm collection of the CREA research centre for Genomics and Bioinformatics (Fiorenzuola d’Arda, Italy), was further propagated in Anhui province, P.R. China (32° 12′ N, 118° 29′ E) in the 2014–2015 growing season. The soil characteristics were evaluated following previous methods (Xue et al. 2016) and are reported in Supplementary Table 1. Genotypes were sown in 1-m rows with three replicates, under a randomized complete block design (Supplementary Fig. 1-A). Seeds harvested from each row were germinated in a vertical germination system as described previously (Xue et al. 2017). The seeds were fixed in each plate with paper slips, thus allowing unimpeded root elongation along the surface of the filter paper (Supplementary Fig. 1-B). Salt and osmotic stress were simulated independently with 80, 160, 240, 320, and 400 mM NaCl, and 5, 10, 15, 20, and 25% PEG6000 solutions, respectively, along with control conditions (no NaCl or PEG6000 added) (Supplementary Fig. 1-B). Normal pure water (with an electrical conductivity of 5.39 μS/cm) used in the control samples and for preparation of salt and PEG6000 solutions was generated by the Mingche Pure/Ultrapure Water System (Millipore).
After 7 days of germination in a growth chamber at 20 °C without light, the filter papers with seedlings were photographed with a fixed camera and the images were analyzed by Image J to measure seminal root and shoot lengths (RL and SL, respectively, Supplementary Fig. 1-C). The entire experiment was replicated three times with seeds from different multiplication plots; experiments under control conditions were run in six replicates.
Mathematical modeling and statistical analysis
Mathematical modeling of seedling growth under multiple salt concentrations was conducted as previously described (Xue et al. 2017) and as illustrated in Supplementary Fig. 1-D, with simulated curves calculated by the Hill function of the Sigmoid model:
The key parameters were extracted from the simulated curves, including Salt-Lmax and Salt80–20. Salt-Lmax represents the maximum seedling lengths in the simulated seedling curves under salt stress, whereas Salt80–20 refers to the salt concentrations leading to 20–80% reduction in seedling Lmax (Xue et al. 2017).
For mathematical modeling of seedling growth under osmotic stress, data of SL and RL were plotted against different concentrations of PEG6000 (0, 5, 10, 15, 20, and 25%), with the control condition assigned to 0% PEG6000 (Supplementary Fig. S1-D). In this case, the 3-parameter Sigmoid function was adopted to derive the simulation curves, since it best fit the six experimental observations with respect to the Hill function:
where variables a, b, and x0 were estimated by Sigmaplot 12.0 and defined as follows: a is a limit value for Lmax, b controls the shape and steepness of the simulated curve, and x0 is the half-maximal activation level of the curve. Lmax was calculated for each genotype by Eq. (2) as follows:
PEG50 represents the PEG6000 concentration on the x-axis leading to half reduction of seedling Lmax and was calculated by solving Eq. (2) as follows:
in the same way, the parameters PEG80, PEG70, PEG60, PEG40, PEG30, and PEG20 were estimated for 80, 70, 60, 40, 30, and 20% reduction of seedling Lmax. To draw consistent plots, values of 1–6 were assigned to the different levels of salt and osmotic stress treatments, as shown in Supplementary Fig. 1-D. Area under the curve (AUC) was calculated as definite integrals by MATLAB 2012a according to the equation of (El-Kassaby et al. 2008):
where 1 and 6 indicate the treatment levels, hence the interval of integrals; f(x) refers to the (1) and (2) function for salt and osmotic stress, respectively. All of the parameters (Salt-Lmax, PEG-Lmax, Salt80-Salt20, PEG80-PEG20, Salt-AUC, and PEG-AUC) extracted from the simulation curves are hereafter termed response parameters (REP). All of the curves were modeled independently from each replicate of every genotype, and REPs were then averaged from three replicates. Under 400 mM NaCl and 25% PEG6000, coleoptiles only emerged from 13 to 12% of all cultivars, and therefore, SL values of 0 were assigned for further data modeling. Overall, 56 phenotypic traits divided into two groups were used for genetic analyses (Table 1); these traits were collected under the three conditions: no stress, salt stress, and osmotic stress. Lmax was regarded as a non-stressed seedling trait because of its strong correlation with seedling length under control conditions (r > 0.9). Repeatability across experiments (H2) was calculated for each trait from the variance components, as previously described (Tondelli et al. 2014):
where σ2e is the residual variance component and σ2g is the genotypic variance component. Principal component analysis (PCA) was performed with all of the phenotypic data collected by using the software PAST (http://folk.uio.no/ohammer/past/) and the results were plotted by Sigmaplot 12.0 (Systat Software, San Jose, CA).
GWA analysis
The European winter barley collection has been genotyped with the Illumina 9K iSelect Chip (Digel et al. 2016), and this allowed detecting row type as the main source of population stratification. In fact, the collection is composed of 65 two-row (2R) and 78 six-row (6R) accessions. In the present work, 4885 gene-based SNP markers were used for GWA analyses, after excluding SNPs with more than 10% of missing data or less than 5% minor allele frequency (MAF). Association analyses were conducted with TASSEL 3.0 (Bradbury et al. 2007) by applying a mixed linear model (MLM) with kinship matrix (K) to control the effects of population structure and genotype relatedness. A threshold p value [−Log10 (p)] = 4 was used to define significant associations. Adjustment of p values for multiple testing was also calculated using the false discovery rate (FDR) method (Benjamini and Hochberg 1995) and a significance level of 0.1 was considered as the FDR threshold in the present work.
Results
Phenotypic characterization of barley seedlings under salt and PEG-mediated osmotic stress
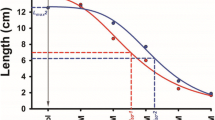
The dynamic responses of barley seedlings to various salt and osmotic stresses are reported in Fig. 1A and B, respectively, for the whole winter barley collection, with representative curves calculated from the mean values of both RL and SL. Similar patterns were observed for RL and SL simulation curves under salt stress (Fig. 1A), but the curves were different under PEG-mediated osmotic stress (Fig. 1B). Under both conditions, seedling length was significantly reduced by the increased stress levels, with the exception of RL under 5% and 10% PEG6000 treatments. As a consequence, a flatter curve was observed for RL under osmotic stress, which clearly differs from the typical S curve observed under salt stress as well as for SL under osmotic stress. The different behaviors of RL under salt and osmotic stress but not SL might indicate a significant ionic effect that influences root more than shoot growth under salt stress.
Dynamic responses of barley seedlings to different levels of salt stress (A) and PEG-mediated osmotic stress (B). Each point represents root or shoot length (RL and SL, respectively) for a single cultivar after 7 days of germination under different growth conditions. Average coefficients of curve fitting are higher than 0.98. Blue and red curves refer to the average values of the whole barley panel for salt and PEG6000 treatments, respectively. Letters on the graph denote statistically significant differences between salt and PEG6000 treatments (Tukey HSD, p < 0.05)
The mean values and data range of the 56 traits collected in the present work are summarized in Supplementary Table 2, while Fig. 2A and B show the observed coefficients of variation (CV) for the whole barley panel. Traits describing seedling response to high levels of stress (e.g., RL-400 and RL-25%) displayed the highest phenotypic variation. In contrast, RL and SL measured under control conditions showed lower CV. As for the REP for salt stress, a gradual reduction of phenotypic variability was observed from RL-Salt80 to RL-Salt20 and from SL-Salt80 to SL-Salt20, whereas there were no clear patterns for REP under PEG-mediated stress. Overall, traits describing the responses to salt stress exhibited higher phenotypic variation (average CV 10%) than those describing responses to PEG-mediated osmotic stress (average CV 9%).
Descriptive statistics of phenotypic traits evaluated in the European winter barley collection. (A and B) Traits are presented in ascending order along the x-axis according to estimated CV. (C and D) Repeatability across experiments (H2) was calculated for all traits. Dashed lines within each graph indicate average values
H2 was calculated for each parameter (Fig. 2C and D), giving values of 0.79 for RL-Control and 0.82 for SL-Control, and average values of 0.69 and 0.67 for traits measured under saline and osmotic conditions, respectively. Under salt stress conditions, the highest H2 values were observed for RL-Salt-AUC, SL-Salt-AUC, and RL-240 mM, while RL-PEG-AUC, RL-PEG-Lmax, and SL-PEG-Lmax exhibited the highest H2 scores under osmotic stress. No clear relationship was observed between CV and H2 during the experiment. Several traits from individual salt and PEG treatments showed higher CV and similar H2 with respect to the REP (e.g., RL-240 mM and RL-25%). Considering individual treatments and REP might lead to different mapping results, we decided to exploit all of the available data for GWA mapping.
GWA mapping of seedling response to salt and PEG-mediated osmotic stress
The 56 phenotypic traits were analyzed by PCA (Supplementary Fig. 2), with the first two components contributing 50.8% of the observed phenotypic variation. Two-row and six-row barley cultivars were not significantly separated by either PC1 or PC2, indicating that row type is not a major determinant for barley seedling growth under the tested conditions. GWA analysis with MLM in TASSEL identified a total of 38 SNPs as significantly associated with 24 traits (considering thresholds of −Log10 (p) ≥ 4.0 and FDR-adjusted p ≤ 0.1). Finally, associated markers were classified into 28 loci based on their physical positions on the barley chromosomes and named QTL1 to QTL28 (Table 2 and Fig. 3A). In total, 15 traits with 19 significant trait–marker associations (−Log10 (p) ≥ 4.0) were excluded by the FDR threshold (p > 0.1); these are listed in Supplementary Table 3.
(A) Genome-wide distribution of QTL detected in the present study. QTL detected under control, salt stress, and PEG-mediated osmotic stress conditions, as well as their corresponding markers, are indicated in green, red, and blue, respectively. Genes related to the analyzed traits are shown in magenta, wherein Fr-H2 gene cluster is highlighted by a gray box. Green boxes indicate overlapping QTL identified under different growth conditions, and blue box refers to the QTL co-localized by RL and SL traits. Physical positions of QTL, markers, and candidate genes are reported according to Mascher et al. (2017). (B) Venn diagrams showing the intersections among QTL. QTL intersections detected under control, salt stress, and PEG-mediated osmotic stress conditions are presented in left, and between root length (RL) and shoot length (SL) traits are listed in right
The highest number of significant associations was detected on chromosomes 1H and 5H, with eight and seven different loci, respectively. Overlap between significant loci was explored by Venn diagram (Fig. 3B) and only four QTL were commonly associated with seedling growth under the different conditions (control, salt stress, and osmotic stress; these QTL are highlighted in green in Fig. 3A), given the rule that multiple traits of each locus associated with identical SNP markers (Table 2). In particular, a genomic region on chromosome 1H (QTL5) was detected in the responses to all three growth conditions. QTL27 (chromosome 6H) was associated with seedling growth under both salt and osmotic stress conditions, whereas two QTL (QTL2 and QTL21) were responsive to both control and PEG treatments. For each of these loci, the SNP allele conferred the same favorable effects under the different growing conditions, as summarized in Supplementary Table 4. Moreover, whereas QTL27 was clearly related to the barley response to stress conditions, QTL2, QTL5, and QTL21 displayed general effects on seedling development. Meanwhile, 8 QTL were specifically associated with seedling growth under saline conditions, while 16 QTL were only detected in response to osmotic stress. Only 1 out of 28 loci (QTL25) was co-localized when comparing QTL for RL and SL traits (Fig. 3B, and highlighted in blue in Fig. 3A), with 21 and 6 loci specifically associated with RL and SL, respectively.
To verify whether known candidate genes map in proximity of the detected QTL, 37 genes (plus Fr-H2 CBF gene cluster) related to seed component (Wang et al. 2015), dormancy (Nakamura et al. 2016), germination (Matthies et al. 2009), seedling growth (Gómez-Cadenas et al. 2001, Chandler and Harding 2013), salt tolerance (Qiu et al. 2011; Rivandi et al. 2011; Shavrukov et al. 2013; Adem et al. 2014; Zhu et al. 2016; Hazzouri et al. 2018), and drought tolerance (Malatrasi et al. 2002) were identified on the reference barley genome sequence of Morex (Mascher et al. 2017) by BLAST analysis (Supplementary Table 5). The flanking markers of a major QTL for barley seedling growth under salt stress previously reported in the OWB population (Witzel et al. 2010), GBS0318 and GBR0227, were also considered in Fig. 3A. Based on these analyses, the positions of HVA1 (Xu et al. 1996) and HvCPI5 (cysteine-proteinase inhibitor) (Martínez et al. 2005) are close to the peak markers of QTL7 and QTL14 on chromosomes 1H and 3H, respectively. The peak marker of QTL26, BOPA1_4771-380, is located 2 Mbp proximal to Fr-H2 locus of HvCBF gene cluster (Francia et al. 2007) on chromosome 5HL. On chromosome 6H, QTL27 is physically linked to four dehydrin protein-encoding genes (HvDHN) (Zhu et al. 2000). None of the ion-homeostasis genes considered here (HvHKTs, HvNHXs, and HvNaxs) mapped close to salt-induced QTL.
QTL23 (475.8 Mbp, 5H), QTL24 (481.7 Mbp, 5H), and QTL25 (504–505 Mbp, 5H) mapped with a large number of salt-responsive traits and were located in genomic regions where QTL for similar traits have been previously identified (Naz et al. 2014; Witzel et al. 2010). They were therefore further characterized (Fig. 4), together with the only locus associated with three different seedling conditions (QTL5, peak marker BOPA1_12492-541 at 482.4 Mbp on chromosome 1H). QTL23 was associated with two RL traits and its peak marker, BOPA2_12_30745, targets the HORVU5Hr1G060810 gene encoding a phosphatidylinositol-4-phosphate 5-kinase (PIP5K). In contrast, two SL traits were mapped to QTL24, with the peak marker BOPA1_65-778 previously detected as a flanking marker for the root trait in the S42IL population (Naz et al. 2014) and not far from the salt-responsive QTL detected in the OWB population at 485.8–487.2 Mbp (flanking markers GBS0318 and GBR0227) (Witzel et al. 2010). Candidate genes in this genomic region include HvDEP1 (dense and erect panicle) (Wendt et al. 2016) near BOPA1_65-778, HvQsd1 (QTL seed dormancy) (Sato et al. 2016) close to GBR0227, as well as the barley ortholog of OsDro1 (deep rooting) (Uga et al. 2013), which is hosted by Contig_134254 from the Morex genome (this gene has yet to be annotated by the latest barley genome of 2017). Beside RL-320 mM, QTL25 is mapped by the same RL traits (RL-Salt20 and RL-Salt30) of QTL23. Meanwhile, a peak marker (BOPA1_1896-1435) associating with SL-320 mM is also co-localized in this region but does not serve as a peak marker for RL traits, hence differs from the four aforementioned overlapping QTL. Nevertheless, this co-localized QTL segment still inherits a possible response to both root and shoot growth. Given this possibility, the putative candidate genes, such as HORVU5Hr1G065950 (cyclic nucleotide gated channel, CNGC) and HORVU5Hr1G066260 (calmodulin-like protein, CML) are annotated (Fig. 4). Finally, genes with putative involvement in general germination and stress response, such as HORVU1Hr1G068760 (Abscisic acid insensitive, ABI) and HORVU1Hr1G068860 (inositol polyphosphate 5-phosphatase, 5PTase), were detected as possible candidates for QTL5 (Fig. 4); candidates of HORVU1Hr1G005090 (protein phosphatase 2C, PP2C) and HORVU5Hr1G011620 (glycosyltransferase, GT) were annotated in QTL2 and QTL21 region, respectively (Fig. 3A).
QTL detected on chromosome 1HL and 5HL. The −Log10 (p) values of marker–trait associations are reported in the right panels. Peak markers of barley QTL and putative candidate genes are indicated in red and magenta, respectively. QTL regions previously reported on 5HL are shown in black. Chromosome regions inheriting the putative candidate genes are depicted in cyan segments
Discussion
Integration of methodologies improves genetic analysis of barley seedling response to salt and osmotic stress
Plant morphological responses to abiotic stresses have been widely investigated to elucidate wilting under drought conditions (Sayed et al. 2012) or chlorosis under salinity (Cui et al. 2015; Fan et al. 2016), and this is one of the most common and convenient approaches to evaluating tolerant phenotypes. Nevertheless, morphological responses of seedlings are the result of dynamic interactions with stresses; hence, both need to be adequately described. Therefore, optimized stress treatments in combination with suitable mathematical modeling are necessary.
One to several concentrations of NaCl or PEG is commonly used to test their effects on seed germination and seedling growth. For example in barley, concentrations of 250, 256, 300, 341, and 427 mM NaCl were used to screen the OWB population (Witzel et al. 2010), SM population (Mano and Takeda 1997), and other RIL populations (Ahmadi-Ochtapeh et al. 2015; Zaare and Jafary 2013), with salt concentrations from 200 to 300 mM NaCl most frequently selected. Consistent with these previous reports, higher CV and H2 values were observed in the present study under treatments with NaCl concentrations of 240 and 320 mM (Fig. 2). With respect to PEG-mediated osmotic stress, PEG6000 (Zhang et al. 2010) and PEG8000 (Ji et al. 2014) are commonly chosen to simulate the osmotic stress imposed by drought, with treatments of 13.6, 15, 18, and 20% PEG6000 having been applied to screen germination and seedling traits in diverse spring barley (Abdel-Ghani et al. 2015), wild barley (Barati et al. 2015), and Tibetan barley (Ahmed et al. 2016) accessions. PEG concentrations below 15% were not used because of low effects, as confirmed by the response curves shown in Fig. 1B. Because of the limited number of treatments, scores/indices are commonly used to define tolerance levels, as well as for QTL mapping for drought (Chen et al. 2010) and salinity (Gimhani et al. 2016). In our study, no significant marker–trait associations were detected from similar indices (data not shown), suggesting that this linear concept might not be applicable to the non-linear trends of seedling growth under a gradient of stress treatments.
In a study of maize seedling behavior under multiple salt stresses, a Sigmoid model fit the growth data better than a linear function (Khan et al. 2003). The dynamic response of seedling growth under a gradient of salt treatments has already been simulated in barley by the Hill function of the Sigmoid model (Xue et al. 2017), and curve-extracted parameters were useful in mapping a major QTL for barley response to salt stress. Similarly, polynomial curves defined by the Sigmoid function largely improved the description of seedling response to multiple osmotic stress treatments, but this has never before been attempted in barley. In our experiments, root response to PEG fit a Sigmoid 3-parameter function better than the Hill equation, and the derived REP resulted in the identification of nine QTL. In general, 18 QTL out of 28 were mapped by REP extracted from both salt and osmotic stress curves, reflecting their strong power. AUC is an additional parameter describing the polynomial curves of seed dormancy (Joosen et al. 2010) which has not been exploited to study abiotic stress tolerance in barley. In the present study, both high H2 and a strong association with SNPs at three QTL suggested it to be a useful parameter for genetic analyses.
Different genomic regions control seedling morphological response to salt and osmotic stress
GWA scans in this European winter barley cultivar collection detected 10 QTL under salt stress and 20 QTL under osmotic stress (Fig. 3B). The difference in QTL numbers between the two stresses was mainly contributed by RL traits (Fig. 3A), consistent with the observation of two different root-response curves (Fig. 1). Only two QTL (QTL5 and QTL27) were detected under both stresses, suggesting distinct genetic control of seedling response, although theoretically, the two stress conditions share osmotic effects; and even, the osmotic effect is considered as the major physiological outcome of exposure to salt during seed germination in barley (Zhang et al. 2010). Patakas (2012) grouped stress-induced morphological responses (SIMR) at the plant anatomical level into three components: inhibition of cell elongation, localized stimulation of cell division, and alterations in cell differentiation status. Since the seeds were only germinated for 7 days in our study, the diverse responses of RL under salt and PEG-mediated osmotic stress probably reflect different abilities to cope with inhibited cell elongation. The ionic effects of salt stress seemed to further inhibit root elongation compared with the PEG-mediated osmotic effect alone, as reflected by the steeper RL curve observed in the salt treatments (Fig. 1).
Comparing the H2 and CV values of 80 mM (0.68, 8%), 160 mM (0.65, 8%), 240 mM (0.78, 19%), 320 mM (0.69, 17%), and 400 mM (0.65, 28%) NaCl with 5% (0.70, 6%), 10% (0.64, 6%), 15% (0.54, 7%), 20% (0.67, 14%), and 25% (0.73, 36%) PEG6000 (Fig. 2), all of the traits had significant trait–marker associations except RL-240 mM and RL-5% which failed to pass significance thresholds (Tables 1 and 2). Although the highest CV was found for RL-25%, on average, RL traits under moderate salt stress (160–320 mM NaCl) showed higher CV relative to osmotic stress (10–20% PEG6000). No clear differences in repeatability (H2) were observed between traits from the two stresses, suggesting that different CV levels were not due to experimental error. This suggests that the ionic effect caused by moderate salt stress enhances the phenotypic variability of RL in the barley collection more than that caused by PEG-mediated stress. The salt stress datasets with such statistical characteristics are expected to achieve more significant loci (comparing with osmotic stress), but in fact, less salt-induced QTL were obtained. Moreover, similar H2 and CV values were obtained for REP of salt and osmotic stresses (Fig. 2). Based on these observations, we conclude that the lower number of salt stress QTLs is not due to the statistical characteristics of the phenotypic data. A possible reason for this genetic discrepancy between the two stresses might be that more genetic variations are inherited by this winter barley panel in osmotic-responsive genes/genetic loci than in salt-induced ones for seedling growth. Nevertheless, several studies have suggested that the SIMR of seedling roots is characterized by growth redistribution rather than cessation (Pasternak 2005; Xue et al. 2017; Zolla et al. 2010). The genetic control of root growth redistribution, such as stimulation of lateral root development, needs to be further investigated.
None of the QTL detected for seedling response to salt stress is associated with one of the previously identified candidate genes for salt stress tolerance at later growth stages (HvHKTs, HvNHXs, HvNaxs, summarized in Supplementary Table 5), which is consistent with other mapping reports (Witzel et al. 2010; Shi et al. 2017). For instance, the best candidate gene for the major QTL detected by Witzel et al. (2010) for seedling growth under salt stress is HvQsd1 (discussed below), a sequence annotated as Alanine aminotransferase encoding gene (Sato et al. 2016). Similar experiments on rice revealed a major QTL, with a nitrate transporter gene being the most plausible candidate for salt tolerance (Shi et al. 2017). Altogether, these evidences suggest that mechanisms relevant for seedling growth under salt stress are distinct from those commonly adopted by plants at the vegetative or adult stages when coping with the same stress.
Stress-non-specific and co-localized QTL for seedling growth and associated candidate genes
Stress-non-specific QTL were first discussed in tomato, where seed germination rate was tested under cold, drought, and saline conditions (Foolad 2003), and QTL promoting rapid germination were found to also improve the germination rate under multiple environmental constraints. Among the four QTL detected under different conditions in the present study, only QTL5 promoted root elongation under the two stress and non-stress growth conditions. In contrast, traits from the two stresses but not from the control condition mapped to QTL27, similar to observations in pearl millet (Sharma et al. 2011). More studies are needed to confirm whether both types of loci can be considered stress-non-specific QTL and be exploited in plant breeding. Finally, no overlapping QTL were mapped for traits scored under control and salt stress, which is consistent with previous results (Xue et al. 2017).
Two QTL regions, QTL2 and QTL21, were only responding to seedling growth in control and PEG-mediated conditions but not the salinities. A candidate gene, PP2C, was annotated in QTL2 region (Fig. 3A), which has been demonstrated with key roles in seed germination (Née et al. 2017) and seedling growth under abiotic stress (Skubacz et al. 2016). A GT gene was noticed at QTL21, reporting with similar ABA signaling at seedling stage (Zhang et al. 2016). It seems that ABA pathways in seedling growth are versatile for osmotic stress.
The peak marker of QTL5 targets the HORVU1Hr1G068760 and HORVU1Hr1G068860 genes encoding ABI like protein and 5PTase, respectively. In Arabidopsis, ABA-responsive element binding protein (AREB) and ABA-responsive element binding factor (ABF) acted as major transcription factors in ABA-responsive gene expression under abiotic stress conditions (Kobayashi et al. 2008). As for barley, HvABI3 (HvVP1) and HvABI5 are the two major AREB/ABFs in ABA regulation during seed germination (Casaretto and Ho 2003). The candidate gene, HORVU1Hr1G068760, belongs to the homolog of HvABI5, which has not been exploited, nevertheless, suggested to be a pleiotropic regulator for seedling growth under multiple conditions in our study. 5PTase constitutes a large protein family that hydrolyzes 5-phosphates from a variety of phosphatidylinositol phosphate and inositol phosphate substrates. It is regulated by abscisic acid, jasmonic acid, and auxin, thus suggesting an important role in signal-transduction pathways (Ercetin and Gillaspy 2004). Its positive role in seedling growth has also been proven in Arabidopsis (Ercetin et al. 2008), as well as a significant role in the regulation of reactive oxygen species and salt tolerance (Kaye et al. 2011). Taken together, the pleiotropic effects make ABI and 5PTase both a suitable candidate gene for QTL5.
The QTL27 peak marker was linked to four different HvDHN genes (Fig. 3A), with HvDHN5 being physically closest. Dehydrins are a group of late embryogenesis-abundant proteins that accumulate during seed development as well as in response to several abiotic stress factors, including low temperature, drought, and salinity (Kosová et al. 2014). Two durum wheat cultivars with contrasting drought tolerance were found to accumulate different levels of DHN5 (Brini et al. 2007), and this pattern was also observed in two cultivars with different salt tolerance (Brini et al. 2007). High expression levels of DHN5 were observed in Tibetan hulless barley under PEG-mediated osmotic, saline, and cold stresses (Du et al. 2011), and this was then confirmed by analyses of barley cultivars under terminal drought stress (Karami et al. 2013). Therefore, we suggest HvDHN5 as a possible candidate gene for QTL27.
As for QTL25, on which RL and SL traits are co-localized, different SNP markers were associated and it was hard to calculate the allele effects on the phenotypes as done in Supplementary Table 4. However, the significant positive correlation between trait SL-320 mM and RL-320 mM (data not shown) indicated that they might favor the same putative allele. As we considered seedling growth for only 7 days, it may control seed germination and subsequently affect both the RL and SL traits. A similar example was reported in a durum wheat RIL population with a QTL on chromosome 4B which had a large impact on both shoot and root traits under no stress conditions (Iannucci et al. 2017), where the semidraft locus Rht-B1 was targeted. In non-stressed barley seedlings, root and shoot traits also co-mapped to a major QTL region and were further linked to the Vrs1 locus on 2H (Wang et al. 2017), but were still distinct from QTL25, which is detected under salt stress only. In terms of the key function of calcium (Ca2+)-dependent signaling under saline conditions (Schulz et al. 2013), two Ca2+-related genes were annotated in this QTL region, CNGC (HORVU5Hr1G065950) and CML (HORVU5Hr1G066260).
CNGC is a Ca2+-permeable cation transport channel that is activated by cyclic nucleotide monophosphates (cNMP) and deactivated by binding Ca2+-calmodulin (CaM) (Tunc-Ozdemir et al. 2013); it was first cloned in the barley aleurone layer as HvCBT1 (Schuurink et al. 1998). Its multiple functions in Arabidopsis seed germination and salinity response have been well summarized (Jha et al. 2016). Less conserved than CaM, CML proteins still serve as Ca2+ sensors in plants and are involved in plant development and multiple abiotic stresses (Bender and Snedden 2013), including seed germination and salt stress. Hence, both candidate genes have potential pleiotropic functions for QTL25. Three RL traits exhibited strong associations with two markers targeting CNGC and CML genes, respectively, and later one also co-mapped by a SL trait (Fig. 4). As regards possible interactions between CNGC and CML proteins, we cannot exclude the possibility that QTL25 is regulated by two underlying genes; this needs further confirmation.
A core QTL region for seedling response to salt stress maps to chromosome 5HL
By comparing the marker–trait associations detected in the present study with previously reported QTL results, a genomic region (QTL24) controlling shoot growth under salt stress was linked to a major QTL mapped in the OWB population on chromosome 5HL (Witzel et al. 2010). The flanking markers GBS0318 and GBR0227 delimited a physical region at 485–487 Mbp, which is 4 Mbp distant from QTL24 (481 Mbp, Fig. 4). One of the two markers, GBS0318, was also detected in the same mapping population as a peak marker for a QTL controlling pre-harvest sprouting (Lohwasser et al. 2013), with the HvQsd1 gene (489 Mbp, Fig. 4) as a suitable candidate (Sato et al. 2016). This gene has been recently cloned and it encodes an alanine aminotransferase protein regulating long-term seed dormancy by catalyzing alanine to glutamate in the seed. Deeper dormancy may result in delayed growth of both shoot and root during germination and seedling establishment, which is compatible with the descriptive scores used in (Witzel et al. 2010), but not with the fact that only SL traits were mapped to QTL24 in our study. It is more similar to the example reported in Arabidopsis, where QTL mapping of early germination under both normal and salt conditions detected the same region on the distal arm of chromosome 4 (Yuan et al. 2016). Therefore, we suggest that the OWB Salt-QTL is independent of the QTL24 detected here. A second major root QTL (S42IL-RI-QTL, 481–484 Mbp, Fig. 4) mapped to the same region, sharing BOPA1_65-778 (481 Mbp) as a flanking marker (Naz et al. 2014). A homologous gene of OsDro1, which is involved in root development in rice, also maps to this QTL region (483 Mbp). OsDro1 is a novel protein with no relevant homology to any other reported proteins, but its roles in controlling root depth and angle suggest that it is a membrane protein (Uga et al. 2013). It seems like a promising candidate for S42IL-RI-QTL but not for QTL24 due to the unmatched traits. Hence, we also suggest that S42IL-RI-QTL is not related to QTL24.
Another gene close to the peak marker of QTL24 is HvDEP1 (482 Mbp). It was first identified as the dense and erect panicle gene in rice, where it significantly improved inflorescence traits and ultimately enhanced crop yield (Huang et al. 2009). In barley, it promotes culm elongation and grain size (Bélanger et al. 2014; Wendt et al. 2016). Its major function in shoot development is consistent with the SL traits mapped to QTL24, despite the different developmental stage. However, no SL traits from the control mapped to this region, probably due to the smaller CV observed for SL in this barley collection. Whether HvDEP1 has a positive function in seedling growth under salt stress needs to be further explored.
Conclusions
In conclusion, 56 different phenotypic traits derived from conditional seedling growth were integrated for GWA mapping and 28 significant loci were detected. Fewer QTL were mapped from salt-stressed seedlings in comparison with osmotically stressed seedlings, and most of them were independent of previously reported loci. At the same time, two types of stress-non-specific QTL inherited pleiotropic effects for seedling growth under different conditions. A co-localized QTL for both root and shoot growth was first exploited from saline conditions. Several candidate genes were highlighted in our study—HvPP2C, HVA1, HvABI, Hv5PTase, HvPIP5K, HvDEP1, HvCNGC, HvCML, and HvDHN5—four related to ABA signaling, two involved in phosphatidylinositol metabolism, and two dependent on Ca2+ signaling. Genes involving in ABA signaling not only suggested as the candidates for osmotically related QTL2, QTL5, and QTL21, but also the QTL7 which was only mapped by osmotic traits. Contrastingly, multiple pathways seemed to be responsive in salt-stressed seedling besides ABA regulations, and not mapped close to known candidate genes (HvHKTs, HvNHXs, and HvNaxs) for salt tolerance at later growth stages, suggesting different mechanisms may be involved. QTL contributing faster growth under non-stress conditions were also identified under salt and osmotic stress, suggesting that the potential dominance of seedling growth benefits its behaviors under abiotic stress. These loci might be exploited in breeding programs to overcome environmental stress at the initial seedling stage.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
Abbreviations
- 5PTase:
-
inositol polyphosphate 5-phosphatase
- ABF :
-
ABA-responsive element binding factor
- ABI :
-
abscisic acid insensitive
- AREB :
-
ABA-responsive element binding protein
- AUC:
-
area under the curve
- CaM:
-
calmodulin
- CML:
-
calmodulin-like protein
- CNGC :
-
cyclic nucleotide gated channel
- CPI :
-
cysteine-proteinase inhibitor
- CV :
-
coefficient of variation
- DEP :
-
dense and erectpanicle
- DHN :
-
dehydrin
- FDR:
-
false discovery rate
- GWA:
-
genome-wide association
- H 2 :
-
repeatability across experiments
- L max :
-
maximum seedling length
- MAF:
-
minor allele frequency
- MLM:
-
mixed linear model
- NT:
-
Nure × Tremois
- OWB:
-
Oregon Wolfe Barley
- PCA:
-
principal component analysis
- PEG:
-
polyethylene glycol
- PP2C :
-
protein phosphatase 2C
- PIP5K :
-
phosphatidylinositol-4-phosphate 5-kinase
- Qsd :
-
QTL seed dormancy
- QTL:
-
quantitative trait loci
- REP:
-
response parameter
- RL:
-
seminal root length
- Salt 50 :
-
salt concentrations leading to 50% reduction of seedling Lmax
- SIMR:
-
stress-induced morphological response
- SL:
-
shoot length
References
Abdel-Ghani AH, Neumann K, Wabila C, Sharma R, Dhanagond S, Owais SJ, Börner A, Graner A, Kilian B (2015) Diversity of germination and seedling traits in a spring barley (Hordeum vulgare L.) collection under drought simulated conditions. Genet Resour Crop Evol 62:275–292. https://doi.org/10.1007/s10722-014-0152-z
Adem GD, Roy SJ, Zhou M, Bowman JP, Shabala S (2014) Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol 14:113. https://doi.org/10.1186/1471-2229-14-113
Ahmadi-Ochtapeh H, Soltanloo H, Ramezanpour SS, Naghavi MR, Nikkhah HR, Rad SY (2015) QTL mapping for salt tolerance in barley at seedling growth stage. Biol Plant 59:283–290. https://doi.org/10.1007/s10535-015-0496-z
Ahmed IM, Nadira UA, Cao F, He X, Zhang G, Wu F (2016) Physiological and molecular analysis on root growth associated with the tolerance to aluminum and drought individual and combined in Tibetan wild and cultivated barley. Planta 243:973–985. https://doi.org/10.1007/s00425-015-2442-x
Bálint AF, Szira F, Börner A, Galiba G (2008) Segregation and association based mapping of loci influencing osmotic tolerance in barley. Acta Biologica Szegediensis 52:101–102
Barati M, Majidi MM, Mirlohi A, Pirnajmodini F, Sharif-Moghaddam N (2015) Response of cultivated and wild barley germplasm to drought stress at different developmental stages. Crop Sci 55:2668–2681. https://doi.org/10.2135/cropsci2015.04.0229
Bayer MM, Rapazote-Flores P, Ganal M, Hedley PE, Macaulay M, Plieske J, Ramsay L, Russell J, Shaw PD, Thomas W, Waugh R (2017) Development and evaluation of a barley 50k iSelect SNP array. Front Plant Sci 8:1792
Bélanger S, Gauthier M, Jean M, Sato K, Belzile F (2014) Genomic characterization of the Hordeum vulgare DEP1 (HvDEP1) gene and its diversity in a collection of barley accessions. Euphytica 198:29–41. https://doi.org/10.1007/s10681-014-1089-1
Bellucci A, Tondelli A, Fangel JU, Torp AM, Xu X, Willats WGT, Flavell A, Cattivelli L, Rasmussen SK (2017) Genome-wide association mapping in winter barley for grain yield and culm cell wall polymer content using the high-throughput CoMPP technique. PLoS One 12:e0173313. https://doi.org/10.1371/journal.pone.0173313
Bender KW, Snedden WA (2013) Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol 163:486–495
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brini F, Hanin M, Lumbreras V, Irar S, Pagès M, Masmoudi K (2007) Functional characterization of DHN-5, a dehydrin showing a differential phosphorylation pattern in two Tunisian durum wheat (Triticum durum Desf.) varieties with marked differences in salt and drought tolerance. Plant Sci 172:20–28. https://doi.org/10.1016/j.plantsci.2006.07.011
Casaretto J, Ho T-hD (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15:271–284
Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Marè C, Tondelli A, Stanca AM (2008) Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crop Res 105:1–14
Chandler PM, Harding CA (2013) ‘Overgrowth’ mutants in barley and wheat: new alleles and phenotypes of the ‘Green Revolution’ DELLA gene. J Exp Bot 64:1603–1613
Chen G, Krugman T, Fahima T, Chen K, Hu Y, Röder M, Nevo E, Korol A (2010) Chromosomal regions controlling seedling drought resistance in Israeli wild barley, Hordeum spontaneum C. Koch. Genet Resour Crop Evol 57:85–99. https://doi.org/10.1007/s10722-009-9453-z
Chloupek O, Hrstková P, Jurecka D, Graner A (2003) Tolerance of barley seed germination to cold- and drought-stress expressed as seed vigour. Plant Breed 122:199–203. https://doi.org/10.1046/j.1439-0523.2003.00800.x
Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, Hedley P, Tondelli A, Pecchioni N, Francia E, Korzun V, Walther A, Waugh R (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44:1388–1392. https://doi.org/10.1038/ng.2447
Cui D, Wu D, Somarathna Y, Xu C, Li S, Li P, Zhang H, Chen H, Zhao L (2015) QTL mapping for salt tolerance based on snp markers at the seedling stage in maize (Zea mays L.). Euphytica 203:273–283. https://doi.org/10.1007/s10681-014-1250-x
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751. https://doi.org/10.1104/pp.106.094532
Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19:371–379. https://doi.org/10.1016/j.tplants.2014.02.001
Digel B, Tavakol E, Verderio G, Tondelli A, Xu X, Cattivelli L, Rossini L, von Korff M (2016) Photoperiod1 (Ppd-H1) controls leaf size. Plant Physiol 172:405–415
Du JB et al (2011) Comparative expression analysis of dehydrins between two barley varieties, wild barley and Tibetan hulless barley associated with different stress resistance. Acta Physiol Plant 33:567–574. https://doi.org/10.1007/s11738-010-0580-0
El-Kassaby YA, Moss I, Kolotelo D, Stoehr M (2008) Seed germination: mathematical representation and parameters extraction. For Sci 54:220–227
Ercetin ME, Gillaspy GE (2004) Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol 135:938–946
Ercetin ME, Ananieva EA, Safaee NM, Torabinejad J, Robinson JY, Gillaspy GE (2008) A phosphatidylinositol phosphate-specific myo-inositol polyphosphate 5-phosphatase required for seedling growth. Plant Mol Biol 67:375–388. https://doi.org/10.1007/s11103-008-9327-3
Fan Y, Zhou G, Shabala S, Chen ZH, Cai S, Li C, Zhou M (2016) Genome-wide association study reveals a new QTL for salinity tolerance in barley (Hordeum vulgare L.). Front Plant Sci 7:946. https://doi.org/10.3389/fpls.2016.00946
Foolad MR (2003) Relationships among cold, salt and drought tolerance during seed germination in tomato: inheritance and QTL mapping. Euphytica 618:47–57
Francia E, Barabaschi D, Tondelli A, Laidò G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115:1083–1091. https://doi.org/10.1007/s00122-007-0634-x
Gimhani DR, Gregorio GB, Kottearachchi NS, Samarasinghe WLG (2016) SNP-based discovery of salinity-tolerant QTLs in a bi-parental population of rice (Oryza sativa). Mol Gen Genomics 291:2081–2099. https://doi.org/10.1007/s00438-016-1241-9
Gómez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13:667–679
Hazzouri KM, Khraiwesh B, Amiri KMA, Pauli D, Blake T, Shahid M, Mullath SK, Nelson D, Mansour AL, Salehi-Ashtiani K, Purugganan M, Masmoudi K (2018) Mapping of HKT1;5 gene in barley using GWAS approach and its implication in salt tolerance mechanism. Front Plant Sci 9. https://doi.org/10.3389/fpls.2018.00156
Honsdorf N, March TJ, Hecht A, Eglinton J, Pillen K (2014) Evaluation of juvenile drought stress tolerance and genotyping by sequencing with wild barley introgression lines. Mol Breed 34:1475–1495. https://doi.org/10.1007/s11032-014-0131-2
Huang X et al (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41:494–497. dx.doi.org/10.1038/ng.352
Iannucci A et al (2017) Mapping QTL for root and shoot morphological traits in a durum wheat × T. dicoccum segregating population at seedling stage. Int J Genomics 2017:6876393. https://doi.org/10.1155/2017/6876393
Jha SK, Sharma M, Pandey GK (2016) Role of cyclic nucleotide gated channels in stress management in plants. Curr Genomics 17:315–329. https://doi.org/10.2174/1389202917666160331202125
Ji H, Liu L, Li K, Xie Q, Wang Z, Zhao X, Li X (2014) PEG-mediated osmotic stress induces premature differentiation of the root apical meristem and outgrowth of lateral roots in wheat. J Exp Bot 65:4863–4872
Joosen RVL, Kodde J, Willems LAJ, Ligterink W, van der Plas LHW, Hilhorst HWM (2010) Germinator: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant J 62:148–159. https://doi.org/10.1111/j.1365-313X.2009.04116.x
Karami A, Shahbazi M, Niknam V, Shobbar ZS, Tafreshi RS, Abedini R, Mabood HE (2013) Expression analysis of dehydrin multigene family across tolerant and susceptible barley (Hordeum vulgare L.) genotypes in response to terminal drought stress. Acta Physiol Plant 35:2289–2297. https://doi.org/10.1007/s11738-013-1266-1
Kaye Y, Golani Y, Singer Y, Leshem Y, Cohen G, Ercetin M, Gillaspy G, Levine A (2011) Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol 157:229–241
Khan AA, Rao SA, McNeilly T (2003) Assessment of salinity tolerance based upon seedling root growth response functions in maize (Zea mays L.). Euphytica 131:81–89. https://doi.org/10.1023/A:1023054706489
Kobayashi F, Maeta E, Terashima A, Takumi S (2008) Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134:74–86. https://doi.org/10.1111/j.1399-3054.2008.01107.x
Kosová K, Vítámvás P, Prášil IT (2014) Wheat and barley dehydrins under cold, drought, and salinity – what can LEA-II proteins tell us about plant stress response? Front Plant Sci 5:343
Lohwasser U, Rehman Arif MA, Börner A (2013) Discovery of loci determining pre-harvest sprouting and dormancy in wheat and barley applying segregation and association mapping. Biol Plant 57:663–674. https://doi.org/10.1007/s10535-013-0332-2
Long NV, Dolstra O, Malosetti M, Kilian B, Graner A, Visser RGF, van der Linden CG (2013) Association mapping of salt tolerance in barley (Hordeum vulgare L.). Theor Appl Genet 126:2335–2351. https://doi.org/10.1007/s00122-013-2139-0
Malatrasi M, Close TJ, Marmiroli N (2002) Identification and mapping of a putative stress response regulator gene in barley. Plant Mol Biol 50:141–150. https://doi.org/10.1023/A:1016051332488
Mano Y, Takeda K (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 94:263–272. https://doi.org/10.1023/A:1002968207362
Martínez M, Abraham Z, Carbonero P, Díaz I (2005) Comparative phylogenetic analysis of cystatin gene families from arabidopsis, rice and barley. Mol Gen Genomics 273:423–432. https://doi.org/10.1007/s00438-005-1147-4
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, Bayer M, Ramsay L, Liu H, Haberer G, Zhang XQ, Zhang Q, Barrero RA, Li L, Taudien S, Groth M, Felder M, Hastie A, Šimková H, Staňková H, Vrána J, Chan S, Muñoz-Amatriaín M, Ounit R, Wanamaker S, Bolser D, Colmsee C, Schmutzer T, Aliyeva-Schnorr L, Grasso S, Tanskanen J, Chailyan A, Sampath D, Heavens D, Clissold L, Cao S, Chapman B, Dai F, Han Y, Li H, Li X, Lin C, McCooke JK, Tan C, Wang P, Wang S, Yin S, Zhou G, Poland JA, Bellgard MI, Borisjuk L, Houben A, Doležel J, Ayling S, Lonardi S, Kersey P, Langridge P, Muehlbauer GJ, Clark MD, Caccamo M, Schulman AH, Mayer KFX, Platzer M, Close TJ, Scholz U, Hansson M, Zhang G, Braumann I, Spannagl M, Li C, Waugh R, Stein N (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544:427–433. https://doi.org/10.1038/nature22043
Matthies IE, Weise S, Röder MS (2009) Association of haplotype diversity in the α-amylase gene amy1 with malting quality parameters in barley. Mol Breed 23:139–152
Mishra AK, Singh VP (2010) A review of drought concepts. J Hydrol 391:202–216. https://doi.org/10.1016/j.jhydrol.2010.07.012
Mondini L, Pagnotta MA (2015) Drought and salt stress in cereals. In: Lichtfouse E, Goyal A (eds) Sustainable agriculture reviews: cereals. Springer International Publishing, Cham, pp 1–31. https://doi.org/10.1007/978-3-319-16988-0_1
Nakamura S et al (2016) Mitogen-activated protein kinase kinase 3 regulates seed dormancy in barley. Curr Biol 26:775–781. https://doi.org/10.1016/j.cub.2016.01.024
Naz AA, Arifuzzaman M, Muzammil S, Pillen K, Léon J (2014) Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genet 15:1–12. https://doi.org/10.1186/s12863-014-0107-6
Née G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ (2017) Delay of germination1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat Commun 8:72. https://doi.org/10.1038/s41467-017-00113-6
Pasternak T (2005) Morphogenic effects of abiotic stress: reorientation of growth in Arabidopsis thaliana seedlings. Environ Exp Bot 53:299–314. https://doi.org/10.1016/j.envexpbot.2004.04.009
Patakas A (2012) Abiotic stress-induced morphological and anatomical changes in plants. In: Ahmad P, Prasad MNV (eds) Abiotic stress responses in plants: metabolism, productivity and sustainability. Springer New York, New York, pp 21–39. https://doi.org/10.1007/978-1-4614-0634-1_2
Qiu L, Wu D, Ali S, Cai S, Dai F, Jin X, Wu F, Zhang G (2011) Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor Appl Genet 122:695–703. https://doi.org/10.1007/s00122-010-1479-2
Rashid A, Hollington PA, Harris D, Khan P (2006) On-farm seed priming for barley on normal, saline and saline–sodic soils in North West Frontier Province, Pakistan. Eur J Agron 24:276–281. https://doi.org/10.1016/j.eja.2005.10.006
Rivandi J, Miyazaki J, Hrmova M, Pallotta M, Tester M, Collins NC (2011) A SOS3 homologue maps to HvNax4, a barley locus controlling an environmentally sensitive Na+ exclusion trait. J Exp Bot 62:1201–1216. https://doi.org/10.1093/jxb/erq346
Rizwan M, Ali S, Ibrahim M, Farid M, Adrees M, Bharwana SA, Zia-ur-Rehman M, Qayyum MF, Abbas F (2015) Mechanisms of silicon-mediated alleviation of drought and salt stress in plants: a review. Environ Sci Pollut Res 22:15416–15431. https://doi.org/10.1007/s11356-015-5305-x
Sato K, Yamane M, Yamaji N, Kanamori H, Tagiri A, Schwerdt JG, Fincher GB, Matsumoto T, Takeda K, Komatsuda T (2016) Alanine aminotransferase controls seed dormancy in barley. Nat Commun 7:11625. https://doi.org/10.1038/ncomms11625
Sayed MA, Schumann H, Pillen K, Naz AA, Léon J (2012) AB-QTL analysis reveals new alleles associated to proline accumulation and leaf wilting under drought stress conditions in barley (Hordeum vulgare L.). BMC Genet 13:61. https://doi.org/10.1186/1471-2156-13-61
Sbei H, Sato K, Shehzad T, Harrabi M, Okuno K (2014) Detection of QTLs for salt tolerance in Asian barley (Hordeum vulgare L.) by association analysis with SNP markers. Breed Sci 64:378–388
Schulz P, Herde M, Romeis T (2013) Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol 163:523–530
Schuurink RC, Shartzer SF, Fath A, Jones RL (1998) Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc Natl Acad Sci 95:1944–1949
Sharma PC, Sehgal D, Singh D, Singh G, Yadav RS (2011) A major terminal drought tolerance QTL of pearl millet is also associated with reduced salt uptake and enhanced growth under salt stress. Mol Breed 27:207–222. https://doi.org/10.1007/s11032-010-9423-3
Sharma PC, Singh D, Sehgal D, Singh G, Hash CT, Yadav RS (2014) Further evidence that a terminal drought tolerance QTL of pearl millet is associated with reduced salt uptake. Environ Exp Bot 102:48–57. https://doi.org/10.1016/j.envexpbot.2014.01.013
Shavrukov Y, Bovill J, Afzal I, Hayes JE, Roy SJ, Tester M, Collins NC (2013) HVP10 encoding V-PPase is a prime candidate for the barley HvNax3 sodium exclusion gene: evidence from fine mapping and expression analysis. Planta 237:1111–1122. https://doi.org/10.1007/s00425-012-1827-3
Shi G, Xia S, Ye J, Huang Y, Liu C, Zhang Z (2015) PEG-simulated drought stress decreases cadmium accumulation in castor bean by altering root morphology. Environ Exp Bot 111:127–134. https://doi.org/10.1016/j.envexpbot.2014.11.008
Shi Y, Gao L, Wu Z, Zhang X, Wang M, Zhang C, Zhang F, Zhou Y, Li Z (2017) Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol 17:92. https://doi.org/10.1186/s12870-017-1044-0
Skubacz A, Daszkowska-Golec A, Szarejko I (2016) The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front Plant Sci 7:1884
Sun C, Gao X, Fu J, Zhou J, Wu X (2015) Metabolic response of maize (Zea mays L.) plants to combined drought and salt stress. Plant Soil 388:99–117. https://doi.org/10.1007/s11104-014-2309-0
Takahashi H, Noda M, Sakurai K, Watanabe A, Akagi H, Sato K, Takeda K (2008) QTLs in barley controlling seedling elongation of deep-sown seeds. Euphytica 164:761–768. https://doi.org/10.1007/s10681-008-9720-7
Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S (2011) OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 62:4863–4874. https://doi.org/10.1093/jxb/err144
Tavakkoli E, Fatehi F, Rengasamy P, McDonald GK (2012) A comparison of hydroponic and soil-based screening methods to identify salt tolerance in the field in barley. J Exp Bot 63:3853–3867
Tondelli A et al (2014) Allelic variation at Fr-H1/Vrn-H1 and Fr-H2 loci is the main determinant of frost tolerance in spring barley. Environ Exp Bot 106:148–155. https://doi.org/10.1016/j.envexpbot.2014.02.014
Tunc-Ozdemir M, Tang C, Ishka MR, Brown E, Groves NR, Myers CT, Rato C, Poulsen LR, McDowell S, Miller G, Mittler R, Harper JF (2013) A cyclic nucleotide-gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiol 161:1010–1020. https://doi.org/10.1104/pp.112.206888
Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45:1097–1102. https://doi.org/10.1038/ng.2725
Varshney RK et al (2012) Genome wide association analyses for drought tolerance related traits in barley (Hordeum vulgare L.). Field Crop Res 126:171–180. https://doi.org/10.1016/j.fcr.2011.10.008
Wang Y, Ren X, Sun D, Sun G (2015) Origin of worldwide cultivated barley revealed by NAM-1 gene and grain protein content. Front Plant Sci 6:803. https://doi.org/10.3389/fpls.2015.00803
Wang Q, Sun G, Ren X, Wang J, Du B, Li C, Sun D (2017) Detection of QTLs for seedling characteristics in barley (Hordeum vulgare L.) grown under hydroponic culture condition. BMC Genet 18:94. https://doi.org/10.1186/s12863-017-0562-y
Wehner GG, Balko CC, Enders MM, Humbeck KK, Ordon FF (2015) Identification of genomic regions involved in tolerance to drought stress and drought stress induced leaf senescence in juvenile barley. BMC Plant Biol 15:125. https://doi.org/10.1186/s12870-015-0524-3
Wendt T, Holme I, Dockter C, Preuß A, Thomas W, Druka A, Waugh R, Hansson M, Braumann I (2016) HvDep1 is a positive regulator of culm elongation and grain size in barley and impacts yield in an environment-dependent manner. PLoS One 11:e0168924. https://doi.org/10.1371/journal.pone.0168924
Witzel K et al (2010) Comparative analysis of the grain proteome fraction in barley genotypes with contrasting salinity tolerance during germination. Plant Cell Environ 33:211–222
Wójcik-Jagła M, Rapacz M, Tyrka M, Kościelniak J, Crissy K, Żmuda K (2013) Comparative QTL analysis of early short-time drought tolerance in Polish fodder and malting spring barleys. Theor Appl Genet 126:3021–3034. https://doi.org/10.1007/s00122-013-2190-x
Xu D, Duan X, Wang B, Hong B, Ho THD, Wu R (1996) Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol 110:249–257
Xue WT, Gianinetti A, Wang R, Zhan ZJ, Yan J, Jiang Y, Fahima T, Zhao G, Cheng JP (2016) Characterizing barley seed macro- and micro-nutrients under multiple environmental conditions. Cereal Res Commun 44:639–649
Xue W, Yan J, Zhao G, Jiang Y, Cheng J, Cattivelli L, Tondelli A (2017) A major QTL on chromosome 7HS controls the response of barley seedling to salt stress in the Nure × Tremois population. BMC Genet 18:79. https://doi.org/10.1186/s12863-017-0545-z
Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y (2012) Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235:253–266. https://doi.org/10.1007/s00425-011-1496-7
Yuan W, Flowers JM, Sahraie DJ, Purugganan MD (2016) Cryptic genetic variation for Arabidopsis thaliana seed germination speed in a novel salt stress environment. G3 (Bethesda) 6:3129
Yun W, Jinping Z, Yong S, Jauhar A, Jianlong X, Zhikang L (2012) Identification of genetic overlaps for salt and drought tolerance using simple sequence repeat markers on an advanced backcross population in rice. Crop Sci 52:1583–1592. https://doi.org/10.2135/cropsci2011.12.0628
Zaare M, Jafary H (2013) Quantitative trait loci diversity for salt tolerance at the early growth stage of barley (Hordeum vulgare L.). Crop Breed J 3:69–77
Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60:3781–3796. https://doi.org/10.1093/jxb/erp214
Zhang H, Irving LJ, McGill C, Matthew C, Zhou D, Kemp P (2010) The effects of salinity and osmotic stress on barley germination rate: sodium as an osmotic regulator. Ann Bot 106:1027–1035
Zhang GZ, Jin SH, Jiang XY, Dong RR, Li P, Li YJ, Hou BK (2016) Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol Biol 90:77–93. https://doi.org/10.1007/s11103-015-0395-x
Zhu B, Choi DW, Fenton R, Close TJ (2000) Expression of the barley dehydrin multigene family and the development of freezing tolerance. Mol Gen Genet 264:145–153. https://doi.org/10.1007/s004380000299
Zhu M, Zhou M, Shabala L, Shabala S (2015) Linking osmotic adjustment and stomatal characteristics with salinity stress tolerance in contrasting barley accessions. Funct Plant Biol 42:252–263
Zhu M, Zhou M, Shabala L, Shabala S (2016) Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ 40:1009–1020. https://doi.org/10.1111/pce.12727
Zhu H, Taylor AA, Astor SR, Terry N (2017) Enhancing saltgrass germination and growth in a saline soil contaminated with petroleum hydrocarbons. Plant Soil 412:189–199. https://doi.org/10.1007/s11104-016-3051-6
Zolla G, Heimer YM, Barak S (2010) Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot 61:211–224. https://doi.org/10.1093/jxb/erp290
Funding
This work was financially supported by a program of the Natural Science Foundation of China (No. 31560578), by China–Israel cooperation program grants from the Ministry of Science and Technology in China (No. 2013DFA32200), and by the FACCE-ERA-NET+ on Climate Smart Agriculture project ClimBar (an integrated approach to evaluating and utilizing genetic diversity for breeding climate-resilient barley).
Author information
Authors and Affiliations
Contributions
AT and LC provided the European barley material, WX designed and performed the experiments, JC, JY, and GZ financed the experiments, ZZ and YJ assisted in the experiments, WX analyzed the data and wrote the manuscript, AT and LC edited the manuscript, and all authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The authors declare that the experiments comply with the current laws of Italy and P. R. of China.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
(DOCX 461 kb)
Supplementary Figure 2
(DOCX 26 kb)
Supplementary Table 1
(DOCX 14 kb)
Supplementary Table 2
(DOCX 19 kb)
Supplementary Table 3
(DOCX 18 kb)
Supplementary Table 4
(DOCX 15 kb)
Supplementary Table 5
(DOCX 22 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Xue, W., Yan, J., Jiang, Y. et al. Genetic dissection of winter barley seedling response to salt and osmotic stress. Mol Breeding 39, 137 (2019). https://doi.org/10.1007/s11032-019-1042-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1042-z