Abstract
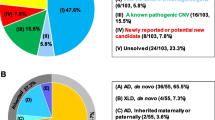
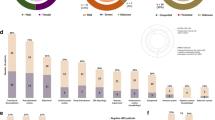
Microcephaly presents heterogeneous genetic etiology linked to several neurodevelopmental disorders (NDD). Copy number variants (CNVs) are a causal mechanism of microcephaly whose investigation is a crucial step for unraveling its molecular basis. Our purpose was to investigate the burden of rare CNVs in microcephalic individuals and to review genes and CNV syndromes associated with microcephaly. We performed chromosomal microarray analysis (CMA) in 185 Brazilian patients with microcephaly and evaluated microcephalic patients carrying < 200 kb CNVs documented in the DECIPHER database. Additionally, we reviewed known genes and CNV syndromes causally linked to microcephaly through the PubMed, OMIM, DECIPHER, and ClinGen databases. Rare clinically relevant CNVs were detected in 39 out of the 185 Brazilian patients investigated by CMA (21%). In 31 among the 60 DECIPHER patients carrying < 200 kb CNVs, at least one known microcephaly gene was observed. Overall, four gene sets implicated in microcephaly were disclosed: known microcephaly genes; genes with supporting evidence of association with microcephaly; known macrocephaly genes; and novel candidates, including OTUD7A, BBC3, CNTN6, and NAA15. In the review, we compiled 957 known microcephaly genes and 58 genomic CNV loci, comprising 13 duplications and 50 deletions, which have already been associated with clinical findings including microcephaly. We reviewed genes and CNV syndromes previously associated with microcephaly, reinforced the high CMA diagnostic yield for this condition, pinpointed novel candidate loci linked to microcephaly deserving further evaluation, and provided a useful resource for future research on the field of neurodevelopment.



Similar content being viewed by others
Data Availability
All the patient’s data reported in this work can be accessed in the DECIPHER website (https://www.deciphergenomics.org/) through the IDs presented in Tables 2 and 4.
References
Abou Jamra, R., Philippe, O., Raas-Rothschild, A., Eck, S. H., Graf, E., Buchert, R., Borck, G., Ekici, A., Brockschmidt, F. F., Nöthen, M. M., Munnich, A., Strom, T. M., Reis, A., & Colleaus, L. (2011). Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. The American Journal of Human Genetics, 88(6), 788–795. https://doi.org/10.1016/j.ajhg.2011.04.019
Abuelo, D. (2007). Microcephaly syndromes. Seminars in Pediatric Neurology, 14, 118–127. https://doi.org/10.1016/j.spen.2007.07.003
Akcakaya, N. H., Capan, Ö. Y., Schulz, H., Sander, T., Caglayan, S. H., & Yapıcı, Z. (2017). De novo 8p23. 1 deletion in a patient with absence epilepsy. Epileptic Disorders, 19(2), 217–221. https://doi.org/10.1684/epd.2017.0906
Almuriekhi, M., Shintani, T., Fahiminiya, S., Fujikawa, A., Kuboyama, K., Takeuchi, Y., Nawaz, Z., Nadaf, J., Kamel, H., Kitam, A. K., Samiha, Z., Mahmoud, L., Ben-Omran, T., Majewski, K., & Noda, M. (2015). Loss-of-function mutation in APC2 causes Sotos syndrome features. Cell Reports, 10(9), 1585–1598. https://doi.org/10.1016/j.celrep.2015.02.011
Asadollahi, R., Oneda, B., Joset, P., Azzarello-Burri, S., Bartholdi, D., Steindl, K., Vincent, M., Cobilanschi, J., Sticht, H., & Baldinger, R. (2014). The clinical significance of small copy number variants in neurodevelopmental disorders. Journal of Medical Genetics, 51(10), 677–688. https://doi.org/10.1136/jmedgenet-2014-102588
Ashwal, S., Michelson, D., Plawner, L., & Dobyns, W. B. (2009). Practice parameter: Evaluation of the child with microcephaly (an evidence-based review): Report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology, 73(11), 887–897. https://doi.org/10.1212/WNL.0b013e3181b783f7
Atli, E. I., Gurkan, H., Atli, E., Vatansever, U., Acunas, B., & Mail, C. (2020). De novo subtelomeric 6p253 deletion with duplication of 6q23. 3–q27: Genotype-phenotype correlation. Journal of Pediatric Genetics, 9(01), 32–39. https://doi.org/10.1055/s-0039-1694703
Ballarati, L., Cereda, A., Caselli, R., Selicorni, A., Recalcati, M. P., Maitz, S., Finelli, P., Larizza, L., & Giardino, D. (2011). Genotype–phenotype correlations in a new case of 8p23. 1 deletion and review of the literature. European Journal of Medical Genetics, 54(1), 55–59. https://doi.org/10.1016/j.ejmg.2010.10.003
Baple, E. L., Maroofian, R., Chioza, B. A., Izadi, M., Cross, H. E., Al-Turki, S., Barwick, K., Skrzypiec, A., Pawlak, R., Wagner, K., Coblentz, R., Zainy, T., Patton, M. A., Mansour, S., Rich, P., Qualmann, B., Hurles, M. E., Kessels, M. M., & Crosby, A. H. (2014). Mutations in KPTN cause macrocephaly, neurodevelopmental delay, and seizures. The American Journal of Human Genetics, 94(1), 87–94. https://doi.org/10.1016/j.ajhg.2013.10.001
Barrie, E. S., Alfaro, M. P., Pfau, R. B., Goff, M. J., McBride, K. L., Manickam, K., & Zmuda, E. J. (2019). De novo loss-of-function variants in NSD2 (WHSC1) associate with a subset of Wolf-Hirschhorn syndrome. Molecular Case Studies, 5(4), a004044. https://doi.org/10.1101/mcs.a004044
Benke, P. J., Hidalgo, R. J., Braffman, B. H., Jans, J., van Gassen, K. L. I., Sunbul, R., & El-Hattab, A. W. (2017). Infantile serine biosynthesis defect due to phosphoglycerate dehydrogenase deficiency: Variability in phenotype and treatment response, novel mutations, and diagnostic challenges. Journal of Child Neurology, 32(6), 543–549. https://doi.org/10.1177/0883073817690094
Berland, S., Haukanes, B. I., Juliusson, P. B., & Houge, G. (2022). Deep exploration of a CDKN1C mutation causing a mixture of Beckwith-Wiedemann and IMAGe syndromes revealed a novel transcript associated with developmental delay. Journal of Medical Genetics, 59(2), 155–164. https://doi.org/10.1136/jmedgenet-2020-107401
Bernardini, L., Radio, F. C., Acquaviva, F., Gorgone, C., Postorivo, D., Torres, B., Alesi, V., Magliozzi, M., Lonardo, F., Monica, M. D., Nardone, A. M., Cesario, C., Mattina, T., Scarano, G., Dallapiccola, B., Digilio, M. C., & Novelli, A. (2018). Small 4p16. 3 deletions: Three additional patients and review of the literature. American Journal of Medical Genetics Part A, 176(11), 2501–2508. https://doi.org/10.1002/ajmg.a.40512
Bernier, R., Golzio, C., Xiong, B., Stessman, H. A., Coe, B. P., Penn, O., Witherspoon, K., Gerdts, J., Baker, C., Vulto-van Silfhout, A. T., Schuurs-Hoeijmakers, J. H., Fichera, M., Bosco, P., Buono, S., Alberti, A., Failla, P., Peeters, H., Steyaert, J., Vissers, L. E. L. M., … & Eichler, E. E. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell, 158(2), 263–276. https://doi.org/10.1016/j.cell.2014.06.017
Bi, W., Cheung, S.-W., Breman, A. M., & Bacino, C. A. (2016). 4p16. 3 microdeletions and microduplications detected by chromosomal microarray analysis: new insights into mechanisms and critical regions. American Journal of Medical Genetics Part A, 170(10), 2540–2550. https://doi.org/10.1002/ajmg.a.37796
Boone, P. M., Campbell, I. M., Baggett, B. C., Soens, Z. T., Rao, M. M., Hixson, P. M., Patel, A., Bi, W., Cheung, S. W., Lalani, S. R., Beaudet, A. L., Stankiewicz, P., Shaw, C. A., & Lupski, J. R. (2013). Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles. Genome Research, 23(9), 1383–1394. https://doi.org/10.1101/gr.156075.113
Boonsawat, P., Joset, P., Steindl, K., Oneda, B., Gogoll, L., Azzarello-Burri, S., Sheth, F., Datar, C., Verma, I. C., & Puri, R. D. (2019). Elucidation of the phenotypic spectrum and genetic landscape in primary and secondary microcephaly. Genetics in Medicine, 21(9), 2043–2058. https://doi.org/10.1038/s41436-019-0464-7
Bruel, A.-L., Franco, B., Duffourd, Y., Thevenon, J., Jego, L., Lopez, E., Deleuze, J.-F., Doummar, D., Giles, R. H., Johnson, C. A., Huynen, M. A., Chevrier, V., Burglen, L., Morleo, M., Desguerres, I., Pierquin, G., Doray, B., Gilbert-Dussardier, B., Reversade, B., … & Thaubin-Robinet, C. (2017). Fifteen years of research on oral–facial–digital syndromes: From 1 to 16 causal genes. Journal of Medical Genetics, 54(6), 371–380 https://doi.org/10.1136/jmedgenet-2016-104436
Canton, A. P., Costa, S. S., Rodrigues, T. C., Bertola, D. R., Malaquias, A. C., Correa, F. A., Arnhold, I. J., Rosenberg, C., & Jorge, A. A. (2014). Genome-wide screening of copy number variants in children born small for gestational age reveals several candidate genes involved in growth pathways. European Journal of Endocrinology, 171(2), 253–262. https://doi.org/10.1530/EJE-14-0232
Capkova, Z., Capkova, P., Srovnal, J., Staffova, K., Becvarova, V., Trkova, M., Adamova, K., Santava, A., Curtisova, V., & Hajduch, M. (2019). Differences in the importance of microcephaly, dysmorphism, and epilepsy in the detection of pathogenic CNVs in ID and ASD patients. PeerJ, 7, e7979. https://doi.org/10.7717/peerj.7979
Carvalho, C. M. B., Vasanth, S., Shinawi, M., Russell, C., Ramocki, M. B., Brown, C. W., Graakjaer, J., Skytte, A.-B., Vianna-Morgante, A. M., & Krepischi, A. C. V. (2014). Dosage changes of a segment at 17p13.1 lead to intellectual disability and microcephaly as a result of complex genetic interaction of multiple genes. The American Journal of Human Genetics, 95(5), 565–578. https://doi.org/10.1016/j.ajhg.2014.10.006
Carvill, G. L., & Mefford, H. C. (2015). CHD2-related neurodevelopmental disorders. In M. P. Adam, D. B. Everman, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. W. Gripp, & A. Amemiya (Eds.), GeneReviews® (pp. 1993–2022). University of Washington.
Catusi, I., Garzo, M., Capra, A. P., Briuglia, S., Baldo, C., Canevini, M. P., Cantone, R., Elia, F., Forzano, F., Galesi, O., Grosso, E., Malacarne, M., Peron, A., Romano, C., Saccani, M., Larizza, L., & Recalcati, M. P. (2021). 8p23. 2-pter microdeletions: Seven new cases narrowing the candidate region and review of the literature. Genes, 12(5), 652. https://doi.org/10.3390/genes12050652
Cheng, H., Dharmadhikari, A. V, Varland, S., Ma, N., Domingo, D., Kleyner, R., Rope, A. F., Yoon, M., Stray-Pedersen, A., Posey, J. E., Crews, S. R., Eldomery, M. K., Akdemir, Z. C., Lewis, A. M., Sutton, V. R., Rosenfeld, J. A., Conboy, E., Agre, K., Xia, F., … & Lyon, G. J.. (2018). Truncating variants in NAA15 are associated with variable levels of intellectual disability, autism spectrum disorder, and congenital anomalies. The American Journal of Human Genetics, 102(5), 985–994. https://doi.org/10.1016/j.ajhg.2018.03.004
Chui, J. V., Weisfeld-Adams, J. D., Tepperberg, J., & Mehta, L. (2011). Clinical and molecular characterization of chromosome 7p22. 1 microduplication detected by array CGH. American Journal of Medical Genetics Part A, 155(10), 2508–2511. https://doi.org/10.1002/ajmg.a.34180
Collins, D. L., & Schimke, R. N. (2011). Regarding cancer predisposition detected by CHG arrays. Genetics in Medicine, 13(11), 982. https://doi.org/10.1097/gim.0b013e31823552a8
Collins, R. L., Glessner, J. T., Porcu, E., Lepamets, M., Brandon, R., Lauricella, C., Han, L., Morley, T., Niestroj, L.-M., Ulirsch, J., Everett, S., Howrigan, D. P., Boone, P. M., Fu, J., Karczewski, K. J., Kellaris, G., Lowther, C., Lucente, D., Mohajeri, K., … & Talkowski, M. E. (2022). A cross-disorder dosage sensitivity map of the human genome. Cell, 185(16), 3041–3055. https://doi.org/10.1016/j.cell.2022.06.036
Cooper, G. M., Coe, B. P., Girirajan, S., Rosenfeld, J. A., Vu, T. H., Baker, C., Williams, C., Stalker, H., Hamid, R., Hannig, V., Abdel-Hamid, H., Bader, P., McCracken, E., Niyazov, D., Leppig, K., These, H., Hummel, M., Alexander, N., Gorski, J., … & Eichler, E. E. (2011). A copy number variation morbidity map of developmental delay. Nature Genetics, 43(9), 838–846. https://doi.org/10.1038/ng.909
Costales, J. L., & Kolevzon, A. (2015). Phelan–McDermid syndrome and SHANK3: Implications for treatment. Neurotherapeutics, 12(3), 620–630. https://doi.org/10.1007/s13311-015-0352-z
der Hagen, M., Pivarcsi, M., Liebe, J., von Bernuth, H., Didonato, N., Hennermann, J. B., Bührer, C., Wieczorek, D., & Kaindl, A. M. (2014). Diagnostic approach to microcephaly in childhood: A two-center study and review of the literature. Developmental Medicine & Child Neurology, 56(8), 732–741. https://doi.org/10.1111/dmcn.12425
DeScipio, C., Conlin, L., Rosenfeld, J., Tepperberg, J., Pasion, R., Patel, A., McDonald, M. T., Aradhya, S., Ho, D., Goldstein, J., McGuire, M., Mulchandani, S., Medne, L., Rupps, R., Serrano, A. H., Thorland, E. C., Tsai, A. C-H, Hilhorst-Hofstee, Y., Ruivenkamp, C. A. L., … & Krantz, I. D. (2012). Subtelomeric deletion of chromosome 10p15. 3: clinical findings and molecular cytogenetic characterization. American Journal of Medical Genetics Part A, 158(9), 2152–2161. https://doi.org/10.1002/ajmg.a.35574
Devhare, P., Meyer, K., Steele, R., Ray, R. B., & Ray, R. (2017). Zika virus infection dysregulates human neural stem cell growth and inhibits differentiation into neuroprogenitor cells. Cell Death & Disease, 8(10), e3106–e3106. https://doi.org/10.1038/cddis.2017.517
Digilio, M. C., Marino, B., Cappa, M., Cambiaso, P., Giannotti, A., & Dallapiccola, B. (2001). Auxological evaluation in patients with DiGeorge/velocardiofacial syndrome (deletion 22q11. 2 syndrome). Genetics in Medicine, 3(1), 30–33. https://doi.org/10.1097/00125817-200101000-00007
Doobin, D. J., Kemal, S., Dantas, T. J., & Vallee, R. B. (2016). Severe NDE1-mediated microcephaly results from neural progenitor cell cycle arrests at multiple specific stages. Nature Communications, 7(1), 1–14. https://doi.org/10.1038/ncomms12551
Dulovic-Mahlow, M., Trinh, J., Kandaswamy, K. K., Braathen, G. J., di Donato, N., Rahikkala, E., Beblo, S., Werber, M., Krajka, V., Busk, Ø. L., Baumann, H., Al-Sannaa, N. A., Hinrichs, F., Affan, R., Navot, N., Al Balwi, M. A., Oprea, G., Holla, Ø. L., Weiss, M. E. R., … & Lohmann, K. (2019). De novo variants in TAOK1 cause neurodevelopmental disorders. The American Journal of Human Genetics, 105(1), 213–220. https://doi.org/10.1016/j.ajhg.2019.05.005
Dworschak, G. C., Draaken, M., Hilger, A. C., Schramm, C., Bartels, E., Schmiedeke, E., Grasshoff-Derr, S., Märzheuser, S., Holland-Cunz, S., Lacher, M., Jenetzky, E., Zwink, N., Schmidt, D., Nöthen, M. M., Ludwig, M., & Reutter, H. (2015). Genome-wide mapping of copy number variations in patients with both anorectal malformations and central nervous system abnormalities. Birth Defects Research Part a: Clinical and Molecular Teratology, 103(4), 235–242. https://doi.org/10.1002/bdra.23321
el Khattabi, L. A., Heide, S., Caberg, J.-H., Andrieux, J., Fenzy, M. D., Vincent-Delorme, C., Callier, P., Chantot-Bastaraud, S., Afenjar, A., Boute-Benejean, O., Cordier, M. P., Faivre, L., Francannet, C., Gerard, M., Goldenberg, A., Masurel-Paulet, A., Mosca-Boidron, A.-L., Marle, N., Moncla, A., … & Pipiras, E. (2020). 16p13.11 microduplication in 45 new patients: refined clinical significance and genotype–phenotype correlations. Journal of Medical Genetics, 57(5), 301–307. https://doi.org/10.1136/jmedgenet-2018-105389
Engels, H., Schüler, H. M., Zink, A. M., Wohlleber, E., Brockschmidt, A., Hoischen, A., Drechsler, M., Lee, J. A., Ludwig, K. U., Kubisch, C., Schwanitz, G., Weber, R. G., Leube, B., Hennekam, R. C. M., Rudnik-Schöneborn, S., Kreiss-Nachtsheim, M., & Reutter, H. (2012). A phenotype map for 14q32. 3 terminal deletions. American Journal of Medical Genetics Part A, 158(4), 695–706. https://doi.org/10.1002/ajmg.a.35256
Enkhmandakh, B., Makeyev, A. V., Erdenechimeg, L., Ruddle, F. H., Chimge, N.-O., Tussie-Luna, M. I., Roy, A. L., & Bayarsaihan, D. (2009). Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. Proceedings of the National Academy of Sciences, 106(1), 181–186. https://doi.org/10.1073/pnas.0811531106
Erdogan, F., Belloso, J. M., Gabau, E., Ajbro, K. D., Guitart, M., Ropers, H. H., Tommerup, N., Ullmann, R., Tümer, Z., & Larsen, L. A. (2008). Fine mapping of a de novo interstitial 10q22–q23 duplication in a patient with congenital heart disease and microcephaly. European Journal of Medical Genetics, 51(1), 81–86. https://doi.org/10.1016/j.ejmg.2007.09.007
Firth, H. V., Richards, S. M., Bevan, A. P., Clayton, S., Corpas, M., Rajan, D., van Vooren, S., Moreau, Y., Pettett, R. M., & Carter, N. P. (2009). DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. The American Journal of Human Genetics, 84(4), 524–533. https://doi.org/10.1016/j.ajhg.2009.03.010
Forzano, F., Napoli, F., Uliana, V., Malacarne, M., Viaggi, C., Bloise, R., Coviello, D., Di Maria, E., Olivieri, I., Di Iorgi, N., & Faravelli, F. (2012). 19q13 microdeletion syndrome: Further refining the critical region. European Journal of Medical Genetics, 55(6–7), 429–432. https://doi.org/10.1016/j.ejmg.2012.03.002
Fritzen, D., Kuechler, A., Grimmel, M., Becker, J., Peters, S., Sturm, M., Hundertmark, H., Schmidt, A., Kreiß, M., Strom, T. M., Wieczorek, D., Haack, T. B., Beck-Wödl, S., Cremer, K., & Engels, H. (2018). De novo FBXO11 mutations are associated with intellectual disability and behavioural anomalies. Human Genetics, 137(5), 401–411. https://doi.org/10.1007/s00439-018-1892-1
Fusco, C., Micale, L., Augello, B., Teresa Pellico, M., Menghini, D., Alfieri, P., Cristina Digilio, M., Mandriani, B., Carella, M., Palumbo, O., Vicari, S., & Merla, G. (2014). Smaller and larger deletions of the Williams Beuren syndrome region implicate genes involved in mild facial phenotype, epilepsy and autistic traits. European Journal of Human Genetics, 22(1), 64–70. https://doi.org/10.1038/ejhg.2013.101
Garret, P., Ebstein, F., Delplancq, G., Dozieres-Puyravel, B., Boughalem, A., Auvin, S., Duffourd, Y., Klafack, S., Zieba, B. A., Mahmoudi, S., Singh, K. K., Duplomb, L., Thauvin-Robinet, C., Costa, J.-M., Krüger, E., Trost, D., Verloes, A., Faivre, L., & Vitobello, A. (2020). Report of the first patient with a homozygous OTUD7A variant responsible for epileptic encephalopathy and related proteasome dysfunction. Clinical Genetics, 97(4), 567–575. https://doi.org/10.1111/cge.13709
Gholkar, A. A., Senese, S., Lo, Y.-C., Vides, E., Contreras, E., Hodara, E., Capri, J., Whitelegge, J. P., & Torres, J. Z. (2016). The X-linked-intellectual-disability-associated ubiquitin ligase Mid2 interacts with astrin and regulates astrin levels to promote cell division. Cell Reports, 14(2), 180–188. https://doi.org/10.1016/j.celrep.2015.12.035
Gilmore, E. C., & Walsh, C. A. (2013). Genetic causes of microcephaly and lessons for neuronal development. Wiley Interdisciplinary Reviews: Developmental Biology, 2(4), 461–478. https://doi.org/10.1002/wdev.89
Girirajan, S., Pizzo, L., Moeschler, J., & Rosenfeld, J. (2015). 16p12.2 recurrent deletion. In M. P. Adam, D. B. Everman, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. W. Gripp, & A. Amemiya (Eds.), GeneReviews® (pp. 1993–2022). University of Washington, Seattle.
Golzio, C., & Katsanis, N. (2013). Genetic architecture of reciprocal CNVs. Current Opinion in Genetics & Development, 23(3), 240–248. https://doi.org/10.1016/j.gde.2013.04.013
Gore, R. H., Nikita, M. E., Newton, P. G., Carter, R. G., Reyes-Bautista, J., & Greene, C. L. (2020). Duplication 6q24: More than just diabetes. Journal of the Endocrine Society, 4(5), bvaa027. https://doi.org/10.1210/jendso/bvaa027
Grünblatt, E., Oneda, B., Ekici, A. B., Ball, J., Geissler, J., Uebe, S., Romanos, M., Rauch, A., & Walitza, S. (2017). High resolution chromosomal microarray analysis in paediatric obsessive-compulsive disorder. BMC Medical Genomics, 10(1), 1–11. https://doi.org/10.1186/s12920-017-0299-5
Hannes, F. D., Sharp, A. J., Mefford, H. C., de Ravel, T., Ruivenkamp, C. A., Breuning, M. H., Fryns, J.-P., Devriendt, K., van Buggenhout, G., Vogels, A., Stewart, H., Hennekam, R. C., Cooper, G. M., Regan, R., Knight, S. J. L., Eichler, E. E., & Vermeesch, J. R. (2009). Recurrent reciprocal deletions and duplications of 16p13. 11: The deletion is a risk factor for MR/MCA while the duplication may be a rare benign variant. Journal of Medical Genetics, 46(4), 223–232. https://doi.org/10.1136/jmg.2007.055202
Helbig, K. L., Lauerer, R. J., Bahr, J. C., Souza, I. A., Myers, C. T., Uysal, B., Schwarz, N., Gandini, M. A., Huang, S., Keren, B., Mignot, C., Afenjar, A., de Villemeur, T. B., Héron, D., Nava, C., Valence, S., Buratti, J., Fagerberg, C. R., Soerensen, K. P., … & Mefford, H. C. (2018). De novo pathogenic variants in CACNA1E cause developmental and epileptic encephalopathy with contractures, macrocephaly, and dyskinesias. The American Journal of Human Genetics, 103(5), 666–678. https://doi.org/10.1016/j.ajhg.2018.09.006
Homma, T. K., Krepischi, A. C. V., Furuya, T. K., Honjo, R. S., Malaquias, A. C., Bertola, D. R., Costa, S. S., Canton, A. P., Roela, R. A., Freire, B. L., Kim, C. A., Rosenberg, C., & Jorge, A. A. L. (2018). Recurrent copy number variants associated with syndromic short stature of unknown cause. Hormone Research in Paediatrics, 89(1), 13–21. https://doi.org/10.1159/000481777
Hotz, A., Hellenbroich, Y., Sperner, J., Linder-Lucht, M., Tacke, U., Walter, C., Caliebe, A., Nagel, I., Saunders, D. E., Wolff, G., Martin, P., & Morris-Rosendahl, D. J. (2013). Microdeletion 5q14. 3 and anomalies of brain development. American Journal of Medical Genetics Part A, 161(9), 2124–2133. https://doi.org/10.1002/ajmg.a.36020
Hu, J., Liao, J., Sathanoori, M., Kochmar, S., Sebastian, J., Yatsenko, S. A., & Surti, U. (2015). CNTN6 copy number variations in 14 patients: A possible candidate gene for neurodevelopmental and neuropsychiatric disorders. Journal of Neurodevelopmental Disorders, 7(1), 1–9. https://doi.org/10.1186/s11689-015-9122-9
Hu, X., Wu, D., Li, Y., Wei, L., Li, X., Qin, M., Li, H., Li, M., Chen, S., Gong, C., & Shen, Y. (2020). The first familial NSD2 cases with a novel variant in a Chinese father and daughter with atypical WHS facial features and a 7.5-year follow-up of growth hormone therapy. BMC Medical Genomics, 13(1), 1–8. https://doi.org/10.1186/s12920-020-00831-9.
Hussain, M. S., Baig, S. M., Neumann, S., Peche, V. S., Szczepanski, S., Nürnberg, G., Tariq, M., Jameel, M., Khan, T. N., Fatima, A., Malik, N. A., Ahmad, I., Altmüller, J., Frommolt, P., Thiele, H., Höhne, W., Yigit, G., Wollnik, B., Neubauer, B. A., … & Noegel, A. A. (2013). CDK6 associates with the centrosome during mitosis and is mutated in a large Pakistani family with primary microcephaly. Human Molecular Genetics, 22(25), 5199–5214. https://doi.org/10.1093/hmg/ddt374
Jacobs, E. Z., Brown, K., Byler, M. C., D’haenens, E., Dheedene, A., Henderson, L. B., Humberson, J. B., van Jaarsveld, R. H., Kanani, F., Lebel, R. R., Millan, F., Oegema, R., Oostra, A., Parker, M. J., Rhodes, L., Saenz, M., Seaver, L. H., Si. Y., Vanlander, A., … & Callewaert, B. (2021). Expanding the molecular spectrum and the neurological phenotype related to CAMTA1 variants. Clinical Genetics, 99(2), 259–268. https://doi.org/10.1111/cge.13874
Jacquinet, A., Brown, L., Sawkins, J., Liu, P., Pugash, D., Van Allen, M. I., & Patel, M. S. (2018). Expanding the FANCO/RAD51C associated phenotype: Cleft lip and palate and lobar holoprosencephaly, two rare findings in Fanconi anemia. European Journal of Medical Genetics, 61(5), 257–261. https://doi.org/10.1016/j.ejmg.2017.12.011
Jansen, S., van der Werf, I. M., Innes, A. M., Afenjar, A., Agrawal, P. B., Anderson, I. J., Atwal, P. S., van Binsbergen, E., van den Boogaard, M.-J., Castiglia, L., Coban-Akdemir, Z. H., van Dijck, A., Doummar, D., van Eerde, A. M., van Essen, A. J., van Gassen, K. L., Sacoto, M. J. G., van Haelst, M. M., Iossifov, I., … & de Vries, B. B. A. (2019). De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. European Journal of Human Genetics, 27(5), 738–746. https://doi.org/10.1038/s41431-018-0292-2
Kalantari, S., & Filges, I. (2020). ‘Kinesinopathies’: Emerging role of the kinesin family member genes in birth defects. Journal of Medical Genetics, 57(12), 797–807. https://doi.org/10.1136/jmedgenet-2019-106769
Kang, S. C., Jaini, R., Hitomi, M., Lee, H., Sarn, N., Thacker, S., & Eng, C. (2020). Decreased nuclear Pten in neural stem cells contributes to deficits in neuronal maturation. Molecular Autism, 11(1), 1–16. https://doi.org/10.1186/s13229-020-00337-2
Kant, S. G., Kriek, M., Walenkamp, M. J. E., Hansson, K. B. M., van Rhijn, A., Clayton-Smith, J., Wit, J. M., & Breuning, M. H. (2007). Tall stature and duplication of the insulin-like growth factor I receptor gene. European Journal of Medical Genetics, 50(1), 1–10. https://doi.org/10.1016/j.ejmg.2006.03.005
Kerzendorfer, C., Hannes, F., Colnaghi, R., Abramowicz, I., Carpenter, G., Vermeesch, J. R., & O’Driscoll, M. (2012). Characterizing the functional consequences of haploinsufficiency of NELF-A (WHSC2) and SLBP identifies novel cellular phenotypes in Wolf-Hirschhorn syndrome. Human Molecular Genetics, 21(10), 2181–2193. https://doi.org/10.1093/hmg/dds033
Kong, Y., Zhou, W., & Sun, Z. (2020). Nuclear receptor corepressors in intellectual disability and autism. Molecular Psychiatry, 25(10), 2220–2236. https://doi.org/10.1038/s41380-020-0667-y
Krepischi, A. C. V., Knijnenburg, J., Bertola, D. R., Kim, C. A., Pearson, P. L., Bijlsma, E., Szuhai, K., Kok, F., Vianna-Morgante, A. M., & Rosenberg, C. (2010). Two distinct regions in 2q24. 2–q24.3 associated with idiopathic epilepsy. Epilepsia, 51(12), 2457–2460. https://doi.org/10.1111/j.1528-1167.2010.02742.x
Krepischi-Santos, A. C. V., Rajan, D., Temple, I. K., Shrubb, V., Crolla, J. A., Huang, S., Beal, S., Otto, P. A., Carter, N. P., & Vianna-Morgante, A. M. (2009). Constitutional haploinsufficiency of tumor suppressor genes in mentally retarded patients with microdeletions in 17p13.1. Cytogenetic and Genome Research, 125(1), 1–7. https://doi.org/10.1159/000218743
Krepischi-Santos, A. C. V., Vianna-Morgante, A. M., Jehee, F. S., Passos-Bueno, M. R., Knijnenburg, J., Szuhai, K., Sloos, W., Mazzeu, J. F., Kok, F., Cheroki, C., Otto, P. A., Mingroni-Netto, R. C., Varela, M., Koiffmann, C., Kim, C. A., Bertola, D. R., Pearson, P. L., & Rosenberg, C. (2006). Whole-genome array-CGH screening in undiagnosed syndromic patients: Old syndromes revisited and new alterations. Cytogenetic and Genome Research, 115(3–4), 254–261. https://doi.org/10.1159/000095922
Leblond, C. S., Nava, C., Polge, A., Gauthier, J., Huguet, G., Lumbroso, S., Giuliano, F., Stordeur, C., Depienne, C., Mouzat, K., Pinto, D., Howe, J., Lemière, N., Durand, C. M., Guibert, J., Ey, E., Toro, R., Peyre, H., Mathieu, A., … & Bourgeron, T. (2014). Meta-analysis of SHANK mutations in autism spectrum disorders: A gradient of severity in cognitive impairments. PLoS Genetics, 10(9), e1004580. https://doi.org/10.1371/journal.pgen.1004580
Lek, M., Karczewski, K. J., Minikel, E. V., Samocha, K. E., Banks, E., Fennell, T., O’Donnell-Luria, A. H., Ware, J. S., Hill, A. J., Cummings, B. B., Tukiainen, T., Birnbaum, D. P., Kosmicki, J. A., Duncan, L. E., Estrada, K., Zhao, F., Zou, J., Pierce-Hoffman, E., Berghout, J., … MacArthur, D. G, & Exome Aggregation Consortium. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536(7616), 285–291. https://doi.org/10.1038/nature19057
Li, L., Yu, Y., Zhang, H., Jiang, Y., Liu, R., & Zhang, H. (2021). Prenatal diagnosis and ultrasonographic findings of partial trisomy of chromosome 6q: A case report and review of the literature. Medicine, 100(2), e24091. https://doi.org/10.1097/MD.0000000000024091
Lim, B. C., Hwang, H., Kim, H., Chae, J.-H., Choi, J., Kim, K. J., Hwang, Y. S., Yum, M.-S., & Ko, T.-S. (2015). Epilepsy phenotype associated with a chromosome 2q24. 3 deletion involving SCN1A: Migrating partial seizures of infancy or atypical Dravet syndrome? Epilepsy Research, 109, 34–39. https://doi.org/10.1016/j.eplepsyres.2014.10.008
Lombardo, B., Esposito, D., Iossa, S., Vitale, A., Verdesca, F., Perrotta, C., di Leo, L., Costa, V., Pastore, L., & Franzé, A. (2019). Intragenic deletion in MACROD2: A family with complex phenotypes including microcephaly, intellectual disability, polydactyly, renal and pancreatic malformations. Cytogenetic and Genome Research, 158(1), 25–31. https://doi.org/10.1159/000499886
Lopes, F., Torres, F., Soares, G., van Karnebeek, C. D., Martins, C., Antunes, D., Silva, J., Muttucomaroe, L., Botelho, L. F., Sousa, S., Rendeiro, P., Tavares, P., Van Esch, H., Rajcan-Separovic, E., & Maciel, P. (2019). The role of AKT3 copy number changes in brain abnormalities and neurodevelopmental disorders: Four new cases and literature review. Frontiers in Genetics, 10, 58. https://doi.org/10.3389/fgene.2019.00058
Loviglio, M. N., Leleu, M., Männik, K., Passeggeri, M., Giannuzzi, G., van der Werf, I., Waszak, S. M., Zazhytska, M., Roberts-Caldeira, I., Gheldof, N., Migliavacca, E., Alfaiz, A. A., Hippolyte, L., Maillard, A. M., 2p15 Consortium, 16p11.2 Consortium, Van Dijck, A., Kooy, R. F., Sanlaville, D., … & Reymond, A. (2017). Chromosomal contacts connect loci associated with autism, BMI and head circumference phenotypes. Molecular Psychiatry, 22(6), 836–849. https://doi.org/10.1038/mp.2016.84
Macé, A., Kutalik, Z., & Valsesia, A. (2018). Copy number variation. In E. Evangelou (Ed.), Genetic epidemiology (pp. 231–258). Springer.
Malvezzi, J. V. M., Magalhaes, H. I., Costa, S. S., Otto, P. A., Rosenberg, C., Bertola, D. R., Fernandes, L. M. W., Vianna-Morgante, A. M., & Krepischi, A. C. V. (2018). KIF11 microdeletion is associated with microcephaly, chorioretinopathy and intellectual disability. Human Genome Variation, 5(1), 1–3. https://doi.org/10.1038/hgv.2018.10
Maulik, P. K., Mascarenhas, M. N., Mathers, C. D., Dua, T., & Saxena, S. (2011). Prevalence of intellectual disability: A meta-analysis of population-based studies. Research in Developmental Disabilities, 32(2), 419–436. https://doi.org/10.1016/j.ridd.2010.12.018
Miller, D. T., Adam, M. P., Aradhya, S., Biesecker, L. G., Brothman, A. R., Carter, N. P., Church, D. M., Crolla, J. A., Eichler, E. E., & Epstein, C. J. (2010). Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. The American Journal of Human Genetics, 86(5), 749–764. https://doi.org/10.1016/j.ajhg.2010.04.006
Mohammadzadeh, A., Akbaroghli, S., Aghaei-Moghadam, E., Mahdieh, N., Badv, R. S., Jamali, P., Kariminejad, R., Chavoshzadeh, Z., Firouzabadi, S. G., Ghanaie, R. M., Nozari, A., Banihashemi, S., Hadipour, F., Hadipour, Z., Kariminejad, A., Najmabadi, H., Shafeghati, Y., & Behjati, F. (2019). Investigation of chromosomal abnormalities and microdeletion/microduplication (s) in fifty Iranian patients with multiple congenital anomalies. Cell Journal, 21(3), 337. https://doi.org/10.22074/cellj.2019.6053
Mohammed, M., Al-Hashmi, N., Al-Rashdi, S., Al-Sukaiti, N., Al-Adawi, K., Al-Riyami, M., & Al-Maawali, A. (2019). Biallelic mutations in AP3D1 cause Hermansky-Pudlak syndrome type 10 associated with immunodeficiency and seizure disorder. European Journal of Medical Genetics, 62(11), 103583. https://doi.org/10.1016/j.ejmg.2018.11.017
Mooneyham, K. A., Holden, K. R., Cathey, S., Dwivedi, A., Dupont, B. R., & Lyons, M. J. (2014). Neurodevelopmental delays and macrocephaly in 17p13. 1 microduplication syndrome. American Journal of Medical Genetics Part A, 164(11), 2887–2891. https://doi.org/10.1002/ajmg.a.36708
Nagamani, S. C. S., Erez, A., Eng, C., Ou, Z., Chinault, C., Workman, L., Coldwell, J., Stankiewicz, P., Patel, A., Lupski, J. R., & Cheung, S. W. (2009). Interstitial deletion of 6q25. 2–q25. 3: A novel microdeletion syndrome associated with microcephaly, developmental delay, dysmorphic features and hearing loss. European Journal of Human Genetics, 17(5), 573–581. https://doi.org/10.1038/ejhg.2008.220
Nevado, J., Rosenfeld, J. A., Mena, R., Palomares-Bralo, M., Vallespín, E., Ángeles Mori, M., Tenorio, J. A., Gripp, K. W., Denenberg, E., & del Campo, M. (2015). PIAS4 is associated with macro/microcephaly in the novel interstitial 19p13.3 microdeletion/microduplication syndrome. European Journal of Human Genetics, 23(12), 1615–1626. https://doi.org/10.1038/ejhg.2015.51
Noor, A., Bogatan, S., Watkins, N., Meschino, W. S., & Stavropoulos, D. J. (2018). Disruption of YWHAE gene at 17p13. 3 causes learning disabilities and brain abnormalities. Clinical Genetics, 93(2), 365–367. https://doi.org/10.1111/cge.13056
Oliveira, D., Leal, G. F., Sertié, A. L., Caires, L. C., Jr., Goulart, E., Musso, C. M., de Oliveira, J. R. M., Krepischi, A. C. V., Vianna-Morgante, A. M., & Zatz, M. (2019). 10q23. 31 microduplication encompassing PTEN decreases mTOR signalling activity and is associated with autosomal dominant primary microcephaly. Journal of Medical Genetics, 56(8), 543–547. https://doi.org/10.1136/jmedgenet-2018-105471
Orellana, C., Roselló, M., Monfort, S., Mayo, S., Oltra, S., & Martinez, F. (2015). Pure duplication of 19p13. 3 in three members of a family with intellectual disability and literature review. Definition of a new microduplication syndrome. American Journal of Medical Genetics Part A, 167(7), 1614–1620. https://doi.org/10.1002/ajmg.a.37046
Palumbo, O., Accadia, M., Palumbo, P., Leone, M. P., Scorrano, A., Palladino, T., Stallone, R., Bonaglia, M. C., & Carella, M. (2018). Refinement of the critical 7p22. 1 deletion region: Haploinsufficiency of ACTB is the cause of the 7p22. 1 microdeletion-related developmental disorders. European Journal of Medical Genetics, 61(5), 248–252. https://doi.org/10.1016/j.ejmg.2017.12.008
Paronett, E. M., Meechan, D. W., Karpinski, B. A., LaMantia, A.-S., & Maynard, T. M. (2015). Ranbp1, deleted in DiGeorge/22q11. 2 deletion syndrome, is a microcephaly gene that selectively disrupts layer 2/3 cortical projection neuron generation. Cerebral Cortex, 25(10), 3977–3993. https://doi.org/10.1093/cercor/bhu285
Parry, D. A., Martin, C.-A., Greene, P., Marsh, J. A., Blyth, M., Cox, H., Donnelly, D., Greenhalgh, L., Greville-Heygate, S., Harrison, V., Lachlan, K., McKenna, C., Quigley, A. J., Rea, G., Robertson, L., Suri, M., & Jackson, A. P. (2021). Heterozygous lamin B1 and lamin B2 variants cause primary microcephaly and define a novel laminopathy. Genetics in Medicine, 23(2), 408–414. https://doi.org/10.1038/s41436-020-00980-3
Pelle, A., Modena, P., Cavallini, A., & Selicorni, A. (2020). Haploinsufficiency of AKT3 gene causing microcephaly and psychomotor delay in a patient with 1q43q44 microdeletion. Clinical Dysmorphology, 29(2), 97–100. https://doi.org/10.1097/MCD.0000000000000313
Pinchefsky, E., Laneuville, L., & Srour, M. (2017). Distal 22q11.2 microduplication: Case report and review of the literature. Child Neurology Open, 4, 2329048X17737651. https://doi.org/10.1177/2329048X17737651
Pivnick, E. K., Qumsiyeh, M. B., Tharapel, A. T., Summitt, J. B., & Wilroy, R. S. (1990). Partial duplication of the long arm of chromosome 6: A clinically recognisable syndrome. Journal of Medical Genetics, 27(8), 523–526. https://doi.org/10.1136/jmg.27.8.523
Quintens, R. (2017). Convergence and divergence between the transcriptional responses to Zika virus infection and prenatal irradiation. Cell Death & Disease, 8(3), e2672. https://doi.org/10.1038/cddis.2017.109
Ramocki, M. B., & Zoghbi, H. Y. (2008). Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature, 455(7215), 912–918. https://doi.org/10.1038/nature07457
Ree, R., Geithus, A. S., Tørring, P. M., Sørensen, K. P., Damkjær, M., Lynch, S. A., & Arnesen, T. (2019). A novel NAA10 p.(R83H) variant with impaired acetyltransferase activity identified in two boys with ID and microcephaly. BMC Medical Genetics, 20(1), 1–8. https://doi.org/10.1186/s12881-019-0803-1
Repnikova, E. A., Lyalin, D. A., McDonald, K., Astbury, C., Hansen-Kiss, E., Cooley, L. D., Pfau, R., Herman, G. E., Pyatt, R. E., & Hickey, S. E. (2020). CNTN6 copy number variations: Uncertain clinical significance in individuals with neurodevelopmental disorders. European Journal of Medical Genetics, 63(1), 103636. https://doi.org/10.1016/j.ejmg.2019.02.008
Rice, A. M., & McLysaght, A. (2017). Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nature Communications, 8(1), 1–11. https://doi.org/10.1038/ncomms14366
Rosenfeld, J. A., Kim, K. H., Angle, B., Troxell, R., Gorski, J. L., Westemeyer, M., Frydman, M., Senturias, Y., Earl, D., Torchia, B., Schultz, R. A., Ellison, J. W., Tsuchiya, K., Zimmerman, S., Smolarek, T. A., Ballif, B. C., & Shaffer, L. G. (2012). Further evidence of contrasting phenotypes caused by reciprocal deletions and duplications: Duplication of NSD1 causes growth retardation and microcephaly. Molecular Syndromology, 3(6), 247–254. https://doi.org/10.1159/000345578
Santiago-Sim, T., Burrage, L. C., Ebstein, F., Tokita, M. J., Miller, M., Bi, W., Braxton, A. A., Rosenfeld, J. A., Shahrour, M., Lehmann, A., Cogné, B., Küry, S., Besnard, T., Isidor, B., Bézieau, S., Hazart, I., Nagakura, H., Immken, L. D. L., Littlejohn, R. O., … & Walkiewicz, M. A. (2017). Biallelic variants in OTUD6B cause an intellectual disability syndrome associated with seizures and dysmorphic features. The American Journal of Human Genetics, 100(4), 676–688. https://doi.org/10.1016/j.ajhg.2017.03.001
Severino, M., Geraldo, A. F., Utz, N., Tortora, D., Pogledic, I., Klonowski, W., Triulzi, F., Arrigoni, F., Mankad, K., Leventer, R. J., Mancini, G. M. S., Barkovich, J. A., Lequin, M. H., & Rossi, A. (2020). Definitions and classification of malformations of cortical development: Practical guidelines. Brain, 143(10), 2874–2894. https://doi.org/10.1093/brain/awaa174
Shaheen, R., Maddirevula, S., Ewida, N., Alsahli, S., Abdel-Salam, G. M. H., Zaki, M. S., Tala, S. AL, Alhashem, A., Softah, A., Al-Owain, M., Alazami, A. M., Abadel, B., Patel, N., Al-Sheddi, T., Alomar, R., Alobeid, E., Ibrahim, N., Hashem, M., Abdulwahab, F., … & Alkuraya, F. S. (2019). Genomic and phenotypic delineation of congenital microcephaly. Genetics in Medicine, 21(3), 545–552. https://doi.org/10.1038/s41436-018-0140-3
Siavrienė, E., Preikšaitienė, E., Maldžienė, Ž, Mikštienė, V., Rančelis, T., Ambrozaitytė, L., Gueneau, L., Reymond, A., & Kučinskas, V. (2020). A de novo 13q31. 3 microduplication encompassing the miR-17∼92 cluster results in features mirroring those associated with Feingold syndrome 2. Gene, 753, 144816. https://doi.org/10.1016/j.gene.2020.144816
Siskos, N., Stylianopoulou, E., Skavdis, G., & Grigoriou, M. E. (2021). Molecular genetics of microcephaly primary hereditary: An overview. Brain Sciences. https://doi.org/10.3390/brainsci11050581
Smajlagić, D., Lavrichenko, K., Berland, S., Helgeland, Ø., Knudsen, G. P., Vaudel, M., Haavik, J., Knappskog, P. M., Njølstad, P. R., & Houge, G. (2021). Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents. European Journal of Human Genetics, 29(1), 205–215. https://doi.org/10.1038/s41431-020-00707-7
Sofou, K., Kollberg, G., Holmström, M., Dávila, M., Darin, N., Gustafsson, C. M., Holme, E., Oldfors, A., Tulinius, M., & Asin-Cayuela, J. (2015). Whole exome sequencing reveals mutations in NARS2 and PARS2, encoding the mitochondrial asparaginyl-tRNA synthetase and prolyl-tRNA synthetase, in patients with Alpers syndrome. Molecular Genetics & Genomic Medicine, 3(1), 59–68. https://doi.org/10.1002/mgg3.115
Soorya, L., Kolevzon, A., Zweifach, J., Lim, T., Dobry, Y., Schwartz, L., Frank, Y., Wang, A. T., Cai, G., Parkhomenko, E., Halpern, D., Grodberg, D., Angarita, B., Willner, J. P., Yang, A., Canitano, R., Chaplin, W., Betancur, C., & Buxbaum, J. D. (2013). Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Molecular Autism, 4(1), 1–17. https://doi.org/10.1186/2040-2392-4-18
Srivastava, S., Love-Nichols, J. A., Dies, K. A., Ledbetter, D. H., Martin, C. L., Chung, W. K., Firth, H. V., Frazier, T., Hansen, R. L., Prock, L., Brunner, H., Hoang, N., Scherer, S. W., Sahin, M., & Miller, D. T. (2019). Meta-analysis and multidisciplinary consensus statement: Exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genetics in Medicine, 21(11), 2413–2421. https://doi.org/10.1038/s41436-019-0554-6
Südhof, T. C. (2017). Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell, 171(4), 745–769. https://doi.org/10.1016/j.cell.2017.10.024
Tang, M., Yang, Y.-F., Xie, L., Chen, J.-L., Zhang, W.-Z., Wang, J., Zhao, T.-L., Yang, J.-F., & Tan, Z.-P. (2015). Duplication of 10q22. 3–q23. 3 encompassing BMPR1A and NGR3 associated with congenital heart disease, microcephaly, and mild intellectual disability. American Journal of Medical Genetics Part A, 167(12), 3174–3179. https://doi.org/10.1002/ajmg.a.37347
Tassano, E., Biancheri, R., Denegri, L., Porta, S., Novara, F., Zuffardi, O., Gimelli, G., & Cuoco, C. (2014). Heterozygous deletion of CHL1 gene: Detailed array-CGH and clinical characterization of a new case and review of the literature. European Journal of Medical Genetics, 57(11–12), 626–629. https://doi.org/10.1016/j.ejmg.2014.09.007
Tolezano, G. C., da Costa, S. S., de Oliveira Scliar, M., Fernandes, W. L. M., Otto, P. A., Bertola, D. R., Rosenberg, C., Vianna-Morgante, A. M., & Krepischi, A. C. V. (2020). Investigating genetic factors contributing to variable expressivity of class I 17p13. 3 microduplication. International Journal of Molecular and Cellular Medicine, 9(4), 296. https://doi.org/10.22088/IJMCM.BUMS.9.4.296
Tsuboyama, M., & Iqbal, M. A. (2021). CHL1 deletion is associated with cognitive and language disabilities–Case report and review of literature. Molecular Genetics & Genomic Medicine, 9(7), e1725. https://doi.org/10.1002/mgg3.1725
Tumiene, B., Čiuladaitė, Ž, Preikšaitienė, E., Mameniškienė, R., Utkus, A., & Kučinskas, V. (2017). Phenotype comparison confirms ZMYND11 as a critical gene for 10p15. 3 microdeletion syndrome. Journal of Applied Genetics, 58(4), 467–474. https://doi.org/10.1007/s13353-017-0408-3
Uddin, M., Unda, B. K., Kwan, V., Holzapfel, N. T., White, S. H., Chalil, L., Woodbury-Smith, M., Ho, K. S., Harward, E., Murtaza, N., Dave, B., Pellecchia, G., D’Abate, L., Nalpathamkalam, T., Lamoureux, S., Wei, J., Speevak, M., Stavropoulos, J., Hope, K. J., … & Singh, K. K. (2018). OTUD7A regulates neurodevelopmental phenotypes in the 15q13. 3 microdeletion syndrome. The American Journal of Human Genetics, 102(2), 278–295. https://doi.org/10.1016/j.ajhg.2018.01.006.
van Woerden, G. M., Bos, M., de Konink, C., Distel, B., Avagliano Trezza, R., Shur, N. E., Barañano, K., Mahida, S., Chassevent, A., Schreiber, A., Erwin, A. L., Gripp, K. W., Rehman, F., Brulleman, S., McCormack, R., de Geus, G., Kalsner, L., Sorlin, A., Bruel, A.-L., … & Kleefstra, T. (2021). TAOK1 is associated with neurodevelopmental disorder and essential for neuronal maturation and cortical development. Human Mutation, 42(4), 445–459. https://doi.org/10.1002/humu.24176
Vargiami, E., Ververi, A., Kyriazi, M., Papathanasiou, E., Gioula, G., Gerou, S., Al-Mutawa, H., Kambouris, M., & Zafeiriou, D. I. (2014). Severe clinical presentation in monozygotic twins with 10p15. 3 microdeletion syndrome. American Journal of Medical Genetics Part A, 164(3), 764–768. https://doi.org/10.1002/ajmg.a.36329
Villela, D., Mazzonetto, P. C., Migliavacca, M. P., Perrone, E., Guida, G., Milanezi, M. F. G., Jorge, A. A. L., Ribeiro-Bicudo, L. A., Kok, F., Campagnari, F., de Rosso-Giuliani, L., da Costa, S. S., Vianna-Morgante, A. M., Pearson, P. L., Krepischi, A. C. V., & Rosenberg, C. (2021). Congenital chromoanagenesis in the routine postnatal chromosomal microarray analyses. American Journal of Medical Genetics Part A, 185(8), 2335–2344. https://doi.org/10.1002/ajmg.a.62237
Vorstman, J., & Scherer, S. W. (2021). What a finding of gene copy number variation can add to the diagnosis of developmental neuropsychiatric disorders. Current Opinion in Genetics & Development, 68, 18–25. https://doi.org/10.1016/j.gde.2020.12.017
Weise, A., Mrasek, K., Klein, E., Mulatinho, M., Llerena, J. C., Jr., Hardekopf, D., Pekova, S., Bhatt, S., Kosyakova, N., & Liehr, T. (2012). Microdeletion and microduplication syndromes. Journal of Histochemistry & Cytochemistry, 60(5), 346–358. https://doi.org/10.1369/0022155412440001
Wentzel, C., Fernström, M., Öhrner, Y., Annerén, G., & Thuresson, A.-C. (2008). Clinical variability of the 22q11.2 duplication syndrome. European Journal of Medical Genetics, 51(6), 501–510. https://doi.org/10.1016/j.ejmg.2008.07.005
Ye, T., Fu, A. K. Y., & Ip, N. Y. (2015). Emerging roles of Axin in cerebral cortical development. Frontiers in Cellular Neuroscience, 9, 217. https://doi.org/10.3389/fncel.2015.00217
Zamboni, V., Jones, R., Umbach, A., Ammoni, A., Passafaro, M., Hirsch, E., & Merlo, G. R. (2018). Rho GTPases in intellectual disability: From genetics to therapeutic opportunities. International Journal of Molecular Sciences, 19(6), 1821. https://doi.org/10.3390/ijms19061821
Zhao, J. J., Halvardson, J., Zander, C. S., Zaghlool, A., Georgii-Hemming, P., Månsson, E., Brandberg, G., Sävmarker, H. E., Frykholm, C., Kuchinskaya, E., Thuresson, A.-C., & Feuk, L. (2018). Exome sequencing reveals NAA15 and PUF60 as candidate genes associated with intellectual disability. American Journal of Medical Genetics Part b: Neuropsychiatric Genetics, 177(1), 10–20. https://doi.org/10.1002/ajmg.b.32574
Zhou, W., He, Y., Rehman, A. U., Kong, Y., Hong, S., Ding, G., Yalamanchili, H. K., Wan, Y.-W., Paul, B., Wang, C., Gong, Y., Zhou, W., Liu, H., Dean, J., Scalais, E., O’Driscoll, M., Morton, J. E. V., DDD study, Hou, X., … & Sun, Z. (2019). Loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus–CA3 projection. Nature Neuroscience, 22(2), 205–217. https://doi.org/10.1038/s41593-018-0311-1
Zhu, W., Li, J., Chen, S., Zhang, J., Vetrini, F., Braxton, A., Eng, C. M., Yang, Y., Xia, F., Keller, K. L., Okinaka-Hu, L., Lee, C., Holder, J. L., Jr., & Bi, W. (2018). Two de novo novel mutations in one SHANK3 allele in a patient with autism and moderate intellectual disability. American Journal of Medical Genetics Part A, 176(4), 973–979. https://doi.org/10.1002/ajmg.a.38622
Zollino, M., Marangi, G., Ponzi, E., Orteschi, D., Ricciardi, S., Lattante, S., Murdolo, M., Battaglia, D., Contaldo, I., Mercuri, E., Stefanini, M. C., Caumes, R., Edery, P., Rossi, M., Piccione, M., Corsello, G., Monica, M. D., Scarano, F., Priolo, M., ... & Zackai, E. (2015). Intragenic KANSL1 mutations and chromosome 17q21. 31 deletions: broadening the clinical spectrum and genotype–phenotype correlations in a large cohort of patients. Journal of Medical Genetics, 52(12), 804–814. https://doi.org/10.1136/jmedgenet-2015-103184
Acknowledgments
The authors would like to thank the patients and their families and the funding agencies São Paulo Research Foundation ([FAPESP] Grant Numbers 2013/080828-1, 2020/15552- 2) and National Council for Scientific and Technological Development ([CNPq] Grant Numbers 157816/2018-4, 305806/2019-0, 140271/2020-1, 303294/2020-5, 303375/2019-1) for supporting this work. This study makes use of data generated by the DECIPHER community. A full list of centres who contributed to the generation of the data is available from https://www.deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome. Those who carried out the original analysis and collection of the Data bear no responsibility for the further analysis or interpretation of them.
Funding
São Paulo Research Foundation ([FAPESP] Grant Numbers 2013/080828-1, 2020/15552-2) and National Council for Scientific and Technological Development ([CNPq] Grant Numbers 157816/2018-4, 305806/2019-0, 140271/2020-1, 303294/2020-5, 303375/2019-1).
Author information
Authors and Affiliations
Contributions
ACVK designed this study and obtained funding; BLF, TKH, RSH, GLY, MRPB, CPK, CAK, AALJ, and DRB referred the patients for CMA; SSC, CR, and ACVK performed the CMA experiments and analysis; GCT and GCB conducted the bibliographic research; AALJ and DRB reviewed the patient's clinical data; GCT wrote the manuscript; GCT, GCB, AMVM, CR, and ACVK reviewed the text. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10803_2022_5853_MOESM1_ESM.xlsx
Supplementary Table 1. List of genes reported to be associated with phenotypes including microcephaly according to OMIM and PubMed. (XLSX 44 kb)
10803_2022_5853_MOESM2_ESM.xlsx
Supplementary Table 2. Full description of the clinical presentation of the 39 Brazilian microcephalic patients carrying pathogenic, likely pathogenic, VUS, and susceptibility CNVs for neurodevelopmental disorders. (XLSX 20 kb)
10803_2022_5853_MOESM3_ESM.jpg
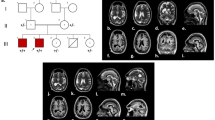
Supplementary Figure 1. Overlapping copy number changes affecting known microcephaly genes that were detected in Brazilian patients and prioritized microcephalic DECIPHER patients carrying <200 kb CNVs. Genomic features of each overlapping segment are displayed with the encompassed protein-coding genes (GENCODE v36 track). Blue line tracks indicate duplications, red lines indicate deletions, and lilac areas highlight known genes for the microcephaly phenotype; darker tracks denote known CNV syndromes already associated with a reduction in brain size. Patients’ DECIPHER IDs are shown; T2 corresponds to “Table 2”, and T4 corresponds to “Table 4” (images derived from the UCSC Genome Browser, up to July 2021). (a) CNVs at 15q26.3 encompassing IGF1R and overlapping with the known chromosome 15q26-qter deletion syndrome region. (b) Overlapping segment at Xq13.1-q13.2, encompassing HDAC8. Note that patient 322306 carries two adjacent CNVs, a 484 kb deletion and a 184 kb duplication. (JPG 418 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tolezano, G.C., Bastos, G.C., da Costa, S.S. et al. Burden of Rare Copy Number Variants in Microcephaly: A Brazilian Cohort of 185 Microcephalic Patients and Review of the Literature. J Autism Dev Disord 54, 1181–1212 (2024). https://doi.org/10.1007/s10803-022-05853-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10803-022-05853-z