Abstract
Background
Microcephaly is a prominent feature of patients with primary autosomal recessive microcephaly 2 (MCPH2) caused by mutations in the WD Repeat Domain 62 (WDR62; OMIM: 613,583).
Aim
The study aimed to identify the underlying genetic factor(s) causing microcephaly in two patients in a consanguineous Iranian family.
Methods
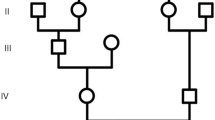
Two male patients (11 and 27 years old) were noticed due to microcephaly, neurodevelopmental delay, and occasional seizures. The younger patient (the proband) was subjected to paired-end whole-exome sequencing followed by Sanger sequencing to detect any underlying genetic factor.
Results
Upon examination, both patients showed microcephaly as a prominent manifestation; they were under-weighted as well. The patients had a moderate gross motor impairment, severe cognitive disability and speech delay, increased deep tendon reflexes, flexible joint contractures, sensorineural hearing loss, and vertical nystagmus as a new ocular finding. The proband had more severe neurodevelopmental delay symptoms. The brain magnetic resonance imaging series revealed severe structural and cortical brain abnormalities in addition to hemiatrophy. Using Whole-exome Sequencing, a novel homozygous missense variant—NM_001083961.2; c.1598A > G: p.(His533Arg)—was identified in the WDR62. Subsequently, in silico analyses determined the possible impacts of the novel variant on the structure and function of WDR62 protein.
Conclusions
Herein, we identified a novel homozygous missense variant in the WDR62 in two patients with MCPH2. Vertical nystagmus and sensorineural hearing loss were detected as novel neurological findings. The present study expands the phenotype and genotype spectrum of MCPH2.



Similar content being viewed by others
Availability of data and materials
The variant was also submitted to ClinVar under the accession code of SCV9076660 and Leiden Open Variation Database (LOVD; individual number: 00331728).
Abbreviations
- WES:
-
Whole-exome sequencing
- MCPH2:
-
Primary autosomal recessive microcephaly 2
- WDR62:
-
WD Repeat Domain 62
- HC:
-
Head circumference
- ACMG/AMP:
-
American College of Medical Genetics and Genomics/Association for Molecular Pathology
- MRI:
-
Magnetic resonance imaging
- MOCA:
-
Montreal Cognitive Assessment
References
Abuelo D (2007) Microcephaly syndromes. Semin Pediatr Neurol 14(3):118–127. https://doi.org/10.1016/j.spen.2007.07.003
Woods CG, Bond J, Enard W (2005) Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet 76(5):717–728
Timothy WY, Mochida GH, Tischfield DJ et al (2010) Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet 42(11):1015–1020
Hofman MA (1984) A biometric analysis of brain size in micrencephalics. J Neurol 231(2):87–93
Barkovich AJ, Kuzniecky RI, Dobyns WB (2001) Radiologic classification of malformations of cortical development. Curr Opin Neurol 14(2):145–149
Razmara E, Azimi H, Tavasoli AR et al (2020) Novel neuroclinical findings of autosomal recessive primary microcephaly 15 in a consanguineous Iranian family. Eur J Med Genet 63(12):104096
Naseer MI, Rasool M, Sogaty S et al (2017) A novel WDR62 mutation causes primary microcephaly in a large consanguineous Saudi family. Ann Saudi Med 37(2):148–153
Bilgüvar K, Oztürk AK, Louvi A et al (2010) Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467(7312):207–210. https://doi.org/10.1038/nature09327
Bhat V, Girimaji S, Mohan G et al (2011) Mutations in WDR62, encoding a centrosomal and nuclear protein. Indian primary microcephaly families with cortical malformations. Clin Genet 80(6):532–540
Roberts E, Jackson AP, Carradice AC et al (1999) The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1–13.2. Eur J Hum Genet 7(7):815–820. https://doi.org/10.1038/sj.ejhg.5200385
Morales A, Kinnamon DD, Jordan E et al (2020) Variant interpretation for dilated cardiomyopathy: refinement of the American College of Medical Genetics and Genomics/ClinGen Guidelines for the DCM Precision Medicine Study. Circ Genom Precis Med 13(4):e002480
Farajollahi Z, Razmara E, Heidari E et al (2020) A novel variant of ST3GAL3 causes non‐syndromic autosomal recessive intellectual disability in Iranian patients. J Genet Med 22(11):e3253
Sepahvand A, Razmara E, Bitarafan F et al (2020) A homozygote variant in the tRNA splicing endonuclease subunit 54 causes pontocerebellar hypoplasia in a consanguineous Iranian family. Mol Genet Genom Med 8(10):e1413
Garshasbi M, Mahmoudi M, Razmara E et al (2020) Identification of RELN variant p.(Ser2486Gly) in an Iranian family with ankylosing spondylitis; the first association of RELN and AS. Eur J Hum Genet 1–9
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol (Clifton, NJ) 132:365–386. https://doi.org/10.1385/1-59259-192-2:365
Wiel L, Baakman C, Gilissen D et al (2019) MetaDome: pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat 40(8):1030–1038
Glaser F, Pupko T, Paz I et al (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19(1):163–164
Frankish A, Harrow J (2014) GENCODE pseudogenes. In Pseudogenes. Springer, pp 129–155
Schwarz JM, Rödelsperger C, Schuelke M, Seelow D (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7(8):575
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31(16):2745–2747
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen‐2. Curr Protoc Hum Genet 76(1):7.20. 21–27.20. 41
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31(13):3812–3814
Sim N-L, Kumar P, Hu J et al (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40(W1):W452–W457
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33(suppl_2):W244-W248
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography 40(1):82–92
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2. 0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33(suppl_2):W306-W310
Rodrigues CH, Pires DE, Ascher DB (2018) DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res 46(W1):W350–W355
Bava KA, Gromiha MM, Uedaira H et al (2004) ProTherm, version 4.0: thermodynamic database for proteins and mutants. Nucleic Acids Res 32(suppl_1):D120-D121
Lim H, Chan T, Yoong T (1994) Denver developmental screening test (DDST). Singapore Med J 35:156–160
Nasreddine ZS, Phillips NA, Bédirian V et al (2005) The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 53(4):695–699
Sherry ST, Ward M-H, Kholodov M et al (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29(1):308–311
Li H, Handsaker B, Wysoker A et al (2009) 1000 Genome project data processing subgroup (2009) The sequence alignment/map format and samtools. Bioinformatics 25(16):2078–2079
Auer PL, Johnsen JM, Johnson AD et al (2012) Imputation of exome sequence variants into population-based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am J Hum Genet 91(5):794–808
Ferrer-Costa C, Gelpí JL, Zamakola L et al (2005) PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21(14):3176–3178
Landrum MJ, Lee JM, Riley GR et al (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42(Database issue):D980–985. https://doi.org/10.1093/nar/gkt1113
Fokkema IF, Taschner PE, Schaafsma GC et al (2011) LOVD v. 2.0: the next generation in gene variant databases. Hum Mutat 32(5):557–563
Nicholas AK, Khurshid M, Désir J et al (2010) WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet 42(11):1010–1014
Bogoyevitch MA, Yeap YY, Qu Z et al (2012) WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J Cell Sci 125(21):5096–5109
Thornton GK, Woods CG (2009) Primary microcephaly: do all roads lead to Rome? Trends Genet 25(11):501–510
Zaqout S, Morris-Rosendahl D, Kaindl AM (2017) Autosomal recessive primary microcephaly (MCPH): an update. Neuropediatrics 48(03):135–142
Barbelanne M, Tsang WY (2014) Molecular and cellular basis of autosomal recessive primary microcephaly. BioMed Res Int
Nicholas AK, Khurshid M, Désir J et al (2010) WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet 42(11):1010–1014
Timothy WY, Mochida GH, Tischfield DJ et al (2010) Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet 42(11):1015–1020
Farag HG, Froehler S, Oexle K et al (2013) Abnormal centrosome and spindle morphology in a patient with autosomal recessive primary microcephaly type 2 due to compound heterozygous WDR62 gene mutation. Orphanet J Rare Dis 8(1):1–14
Faheem M, Naseer MI, Rasool M et al (2015) Molecular genetics of human primary microcephaly: an overview. BMC Med Genomics 8(1):1–11
Ruaud L, Drunat S, Elmaleh-Bergès M et al ( 2021) Neurological outcome in WDR62 primary microcephaly. Dev Med Child Neurol (DMCN) n/a (n/a). https://doi.org/10.1111/dmcn.15060
Naseer MI, Rasool M, Sogaty S et al (2017) A novel WDR62 mutation causes primary microcephaly in a large consanguineous Saudi family. Ann Saudi Med 37(2):148–153
Slezak R, Smigiel R, Obersztyn E et al (2021) Further delineation of phenotype and genotype of primary microcephaly syndrome with cortical malformations associated with mutations in the WDR62 gene. Genes 12(4). https://doi.org/10.3390/genes12040594
Banerjee S, Chen H, Huang H et al (2016) Novel mutations c. 28G > T (p. Ala10Ser) and c. 189G > T (p. Glu63Asp) in WDR62 associated with early onset acanthosis and hyperkeratosis in a patient with autosomal recessive microcephaly type 2. Oncotarget 7(48):78363
Acknowledgements
We are grateful to the family for their willing participation and cooperation with us.
Author information
Authors and Affiliations
Contributions
Hajar Aryan: conceptualization, data curation, formal analysis, methodology, validation. Shaghayegh Zokaei: formal analysis, methodology, validation, visualization, software, writing — original draft, writing — review and editing. Dariush Farhud: data curation, formal analysis. Mohammad Keykhaei: data curation, writing — review and editing. Mahmoud Reza Ashrafi: formal analysis. Maryam Rasulinezhad: formal analysis, validation. Seyyed Mohammad Mahdi Hosseini: formal analysis. Ehsan Razmara: formal analysis, validation, visualization, software, writing — original draft, writing — review and editing. Ali Reza Tavasoli: funding acquisition, resources, methodology, project administration, investigation, writing — review and editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethics committee of Tarbiat Modares University, Tehran, Iran and has therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Consent for publication
Written informed consent was obtained from the patient and patient’s legal guardian for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aryan, H., Zokaei, S., Farhud, D. et al. Novel phenotype and genotype spectrum of WDR62 in two patients with associated primary autosomal recessive microcephaly. Ir J Med Sci 191, 2733–2741 (2022). https://doi.org/10.1007/s11845-021-02890-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-021-02890-y