Abstract
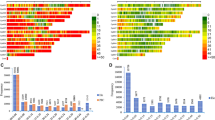
Germplasm collections of major crops include thousands of accessions whose agronomic evaluation is prevented by high costs, while their molecular characterization is getting increasingly affordable. This study aimed to assess the consistency between alfalfa (Medicago sativa L.) landrace diversity measures relative to adaptation pattern, eleven morphophysiological traits, and four genotyping strategies using SSR or genotyping-by-sequencing (GBS) SNP markers recorded on either five individual plants or three bulks of 100 plants each per accession. We focused on eleven landraces from Northern Italy featuring large genotype × environment interaction for biomass yield across environments with contrasting level of summer drought. We recorded 34 SSR and 237 SNP polymorphic markers for individual plants, and 41 SSR and 1274 SNP markers for bulked plants. Landrace diversity based on SNP data from bulked plants was the only molecular diversity measure associated with both adaptive diversity as expressed by pattern analysis (r = 0.34) and drought stress at collecting sites (r = 0.50). No molecular diversity measure was associated with overall morphophysiological diversity. Molecular diversity was highly consistent only between SSR and SNP data from bulked plants (r = 0.67). SSR-based diversity from individual plants exhibited marked inconsistency with other molecular diversity measures but was useful for genetic structure assessment, indicating much smaller among-population variation relative to within-population variation (7.5% in terms of variance ratio; 7.8% as global FST). Moderate inferences on germplasm adaptive diversity may be achieved by molecular characterization that maximizes the sampling of genomic areas and genotypes within accession through GBS applied to bulked plants.


Similar content being viewed by others
References
Annicchiarico P (1992) Cultivar adaptation and recommendation from alfalfa trials in Northern Italy. J Genet Breed 46:269–278
Annicchiarico P (2004) A low-cost procedure for multi-purpose, large-scale field evaluation of forage crop genetic resources. Euphytica 140:223–229
Annicchiarico P (2006a) Diversity, genetic structure, distinctness and agronomic value of Italian lucerne (Medicago sativa L.) landraces. Euphytica 148:269–282
Annicchiarico P (2006b) Prediction of indirect selection for lucerne seed and forage yield from space-planted evaluation. Plant Breed 125:641–643
Annicchiarico P (2007a) Wide- versus specific-adaptation strategy for lucerne breeding in northern Italy. Theor Appl Genet 114:647–657
Annicchiarico P (2007b) Lucerne shoot and root traits associated with adaptation to favourable or drought-stress environments and to contrasting soil types. Field Crops Res 102:51–59
Annicchiarico P, Carelli M (2014) Origin of Ladino white clover as inferred from patterns of molecular and morphophysiological diversity. Crop Sci 54:2696–2706
Annicchiarico P, Piano E (2005) Use of artificial environments to reproduce and exploit genotype × location interaction for lucerne in northern Italy. Theor Appl Genet 110:219–227
Annicchiarico P, Royo C, Bellah F, Moragues M (2009) Relationships among adaptation patterns, morphophysiological traits and molecular markers in durum wheat. Plant Breed 128:164–171
Annicchiarico P, Pecetti L, Abdelguerfi A, Bouizgaren A, Carroni AM, Hayek T, M’Hammadi Bouzina M, Mezni M (2011) Adaptation of landrace and variety germplasm and selection strategies for lucerne in the Mediterranean basin. Field Crops Res 120:283–291
Annicchiarico P, Barrett B, Brummer EC, Julier B, Marshall AH (2015a) Achievements and challenges in improving temperate perennial forage legumes. Crit Rev Plant Sci 34:327–380
Annicchiarico P, Nazzicari N, Li X, Wei Y, Pecetti L, Brummer EC (2015b) Accuracy of genomic selection for alfalfa biomass yield in different reference populations. BMC Genom 16:1020
Annicchiarico P, Nazzicari N, Ananta A, Carelli M, Wei Y, Brummer EC (2016) Assessment of cultivar distinctness in alfalfa: a comparison of genotyping-by-sequencing, SSR marker and morphophysiological observations. Plant Genome 9:2
Annicchiarico P, Bottazzi P, Ruozzi F, Russi L, Pecetti L (2020) Lucerne cultivar adaptation to Italian geographic areas is affected crucially by the selection environment and encourages the breeding for specific adaptation. Euphytica 216:50
Arnold B, Corbett-Detig RB, Hartl D, Bomblies K (2013) RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol 22:3179–3190
Biazzi E, Nazzicari N, Pecetti L, Brummer EC, Palmonari A, Tava A, Annicchiarico P (2017) Genome-wide association mapping and genomic selection for alfalfa (Medicago sativa) forage quality traits. PLoS ONE 12:e0169234
Bolaños-Aguilar ED, Huyghe C, Julier B, Ecalle C (2000) Genetic variation for seed yield and its components in alfalfa (Medicago sativa L.) populations. Agronomie 20:333–345
Charcosset A, Moreau L (2004) Use of molecular markers for the development of new cultivars and the evaluation of genetic diversity. Euphytica 137:81–94
Clark LV, Schreier AD (2017) Resolving microsatellite genotype ambiguity in populations of allopolyploid and diploidized autopolyploid organisms using negative correlations between allelic variables. Mol Ecol Resour 17:1090–1103
Crochemore ML, Huyghe C, Kerlan MC, Durand F, Julier B (1996) Partitioning and distribution of RAPD variation in a set of populations of the Medicago sativa complex. Agronomie 16:421–432
Crochemore ML, Huyghe C, Écalle C, Julier B (1998) Structuration of alfalfa genetic diversity using agronomic and morphological characteristics. Relationship with RAPD markers. Agronomie 18:79–94
De Silva HN, Hall AJ, Rikkerink E, Fraser LG (2005) Estimation of allele frequencies in polyploids under certain patterns of inheritance. Heredity 95:327–334
DeLacy IH, Basford KE, Cooper M, Bull IK, McLaren CG (1996) Analysis of multi-environment trials—an historical perspective. In: Cooper M, Hammer GL (eds) Plant adaptation and crop improvement. CAB International, Wallingford, pp 39–124
Dias PMB, Julier B, Sampoux JP, Barre P, Dall’Agnol M (2008) Genetic diversity in red clover (Trifolium pratense L.) revealed by morphological and microsatellite (SSR) markers. Euphytica 160:189–205
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379
Ergon Å, Skøt L, Sæther VE, Rognli OA (2019) Allele frequency changes provide evidence for selection and identification of candidate loci for survival in red clover (Trifolium pratense L.). Front Plant Sci 10:718
Estoup A, Jarne P, Cornuet JM (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol 11:1591–1604
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Gauch HG (1992) Statistical analysis of regional yield trials: AMMI analysis of factorial designs. Elsevier, Amsterdam
George J, Dobrowolski MP, Van Zijll De Jong E, Cogan NOI, Smith KF, Forster JW (2006) Assessment of genetic diversity in cultivars of white clover (Trifolium repens L.) detected by SSR polymorphisms. Genome 49:919–930
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9(2):e90346
Goudet J, Jombart T (2015) hierfstat: estimation and tests of hierarchical F-statistics. R package version 0.04-22. 2015. https://CRAN.R-project.org/package=hierfstat
Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338
Greene SL, Gritsenko M, Vandemark G (2004) Relating morphologic and RAPD marker variation to collection site environment in wild populations of red clover (Trifolium pratense L.). Genet Resour Crop Evol 51:643–653
Guichoux E, Lagache L, Wagner S, Chaumeil P, Léger P, Lepais O, Lepoittevin C, Malausa T, Revardel E, Salin F, Petit R (2011) Current trends in microsatellite genotyping. Mol Ecol Resour 11:591–611
Hakl J, Mofidian SMA, Kozová Z, Fuksa P, Jaromír Š (2019) Estimation of lucerne yield stability for enabling effective cultivar selection under rainfed conditions. Grass Forage Sci 74:687–695
Hamblin MT, Warburton ML, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS ONE 2:e1367
Haussmann BIG, Parzies HK, Presterl T, Sušić Z, Miedaner T (2004) Plant genetic resources and crop improvement. Plant Genet Res 2:3–21
Herrmann D, Flajoulot S, Barre P, Huyghe C, Ronfort J, Julier B (2018) Comparison of morphological traits and molecular markers to analyse diversity and structure of alfalfa (Medicago sativa L.) cultivars. Genet Res Crop Evol 65:527
IRRI (2009) Cropstat version 7.2. International Rice Research Institute, Manila
Jia C, Zhao F, Wang X, Han J, Zhao H, Liu G, Wang Z (2018) Genomic prediction for 25 agronomic and quality traits in alfalfa (Medicago sativa). Front Plant Sci 9:1220
Julier B, Huyghe C, Ecalle C (2000) Within- and among-cultivar genetic variation in alfalfa: forage quality, morphology, and yield. Crop Sci 40:365–369
Julier B, Barre P, Lambroni P, Delaunay S, Thomasset M, Lafaillette F, Gensollen V (2018) Use of GBS markers to distinguish among lucerne varieties, with comparison to morphological traits. Mol Breed 38:133
Kölliker R, Jones ES, Jahufer MZZ, Forster JW (2001) Bulked AFLP analysis for the assessment of genetic diversity in white clover (Trifolium repens L.). Euphytica 121:305–315
Li X, Wei Y, Acharya A, Hansen JL, Crawford JL, Viands DR, Michaud R, Claessens A, Brummer EC (2015) Genomic prediction of biomass yield in two selection cycles of a tetraploid alfalfa breeding population. Plant Genome 8:2
Linhart YB, Grant MC (1996) Evolutionary significance of local genetic differentiation in plants. Annu Rev Ecol Syst 27:237–277
Liu S, Feuerstein U, Luesink W, Schultze S, Asp T, Studer B, Becker HC, Dehmer KJ (2018) DArT, SNP, and SSR analyses of genetic diversity in Lolium perenne L. using bulk sampling. BMC Genet 19:10
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Muller MH, Poncet C, Prosperi JM, Santoni S, Ronfort J (2006) Domestication history in the Medicago sativa species complex: inferences from nuclear sequence polymorphism. Mol Ecol 15:1589–1602
Nazzicari N, Biscarini F, Cozzi P, Brummer EC, Annicchiarico P (2016) Marker imputation efficiency for genotyping-by-sequencing data in rice (Oryza sativa) and alfalfa (Medicago sativa). Mol Breed 36:69
Pagnotta MA, Annicchiarico P, Farina A, Proietti S (2011) Characterizing the molecular and morphophysiological diversity of Italian red clover. Euphytica 179:393–404
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420
Pembleton KG, Smith RS, Rawnsley RP, Donaghy DJ, Humphries AW (2010) Genotype by environment interactions of lucerne (Medicago sativa L.) in a cool temperate climate. Crop Pasture Sci 61:493–502
Prosperi JM, Auricht Génier G, Johnson R (2001) Medics (Medicago L.). In: Maxted N, Bennett SJ (eds) Plant genetic resources of legumes in the Mediterranean. Kluwer Academic Publishers, Dordrecht, pp 99–114
Pupilli F, Businelli S, Paolocci F, Scotti C, Damiani F, Arcioni S (1996) Extent of RFLP variability in tetraploid populations of alfalfa, Medicago sativa. Plant Breed 115:106–112
Qiang H, Chen Z, Zhang Z, Wang X, Gao H, Wang Z (2015) Molecular diversity and population structure of a worldwide collection of cultivated tetraploid alfalfa (Medicago sativa subsp. sativa L.) germplasm as revealed by microsatellite markers. PLoS ONE 10:e0124592
Riday H, Reisen P, Raasch JA, Santa-Martinez E, Brunet J (2015) Selfing rate in an alfalfa seed production field pollinated with leafcutter bees. Crop Sci 55:1087
Roldán-Ruiz I, van Eeuwijk FA, Gilliland TJ, Dubreuil P, Dillmann C, Lallemand J, De Loose M, Baril CP (2001) A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium perenne L.) varieties. Theor Appl Genet 103:1138–1150
Şakiroǧlu M, Doyle JJ, Brummer EC (2010) Inferring population structure and genetic diversity of broad range of wild diploid alfalfa (Medicago sativa L.) accessions using SSR markers. Theor Appl Genet 121:403–415
Şakiroǧlu M, Moore KJ, Brummer EC (2011) Variation in biomass yield, cell wall components, and agronomic traits in a broad range of diploid alfalfa accessions. Crop Sci 51:1956–1964
Smýkal P, Coyne CJ, Ambrose MJ, Maxted N, Shaefer H, Blair MW, Berger J, Greene SL, Nelson MN, Besharat N, Vymyslický T, Toker C, Saxena RK, Roorkiwal M, Pandey MK, Hu J, Li YH, Wang LX, Guo Y, Qiu LJ, Redden RJ, Varshney RK (2015) Legume crops phylogeny and genetic diversity for science and breeding. Crit Rev Plant Sci 34:43–104
UPOV (2013) Guidance of the use of biochemical and molecular markers in the examination of distinctness, uniformity and stability (DUS). Document TGP/15. UPOV, Geneva
van Brummelen G (2012) Heavenly mathematics: the forgotten art of spherical trigonometry. Princeton University Press, Princeton
Verwimp C, Ruttink T, Muylle H, Van Glabeke S, Cnops G, Quataert P, Honnay O, Roldàn-Ruiz I (2018) Temporal changes in genetic diversity and forage yield of perennial ryegrass in monoculture and in combination with red clover in swards. PLoS ONE 13:e0206571
Acknowledgements
We are grateful to Y. Wei for her contribution to generation of SNP marker data and to P. Gaudenzi for technical assistance in SSR marker genotyping.
Funding
This research was funded by the Italian Ministry of Agricultural, Food and Forestry Policies within the project Plant Genetic resources/FAO Treaty and by the Samuel Roberts Noble Foundation.
Author information
Authors and Affiliations
Contributions
Conceptualization and funding acquisition, PA and ECB; data generation, PA, MC and NN; data analysis, NN and PA; draft preparation, PA. All authors reviewed and commented on the manuscript and approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Availability of data and materials
Molecular and morphophysiological data are provided as Online Resource 2.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Name, origin, linkage group, locus, observed size range, and numbers of total polymorphic alleles, for 41 SSR markers used for bulked plants and 34 SSR markers used for analysis of individual genotypes of 11 alfalfa landraces (PDF 23 kb)
Online Resource 2
Molecular and morphophysiological data of 11 alfalfa landraces (XLS 540 kb)
Rights and permissions
About this article
Cite this article
Annicchiarico, P., Brummer, E.C., Carelli, M. et al. Strategies of molecular diversity assessment to infer morphophysiological and adaptive diversity of germplasm accessions: an alfalfa case study. Euphytica 216, 98 (2020). https://doi.org/10.1007/s10681-020-02637-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-020-02637-3