Abstract
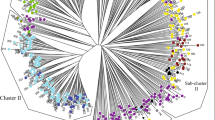
Diversity analyses in alfalfa have mainly evaluated genetic relationships of cultivated germplasm, with little known about variation in diploid germplasm in the M. sativa–falcata complex. A collection of 374 individual genotypes derived from 120 unimproved diploid accessions from the National Plant Germplasm System, including M. sativa subsp. caerulea, falcata, and hemicycla, were evaluated with 89 polymorphic SSR loci in order to estimate genetic diversity, infer the genetic bases of current morphology-based taxonomy, and determine population structure. Diploid alfalfa is highly variable. A model-based clustering analysis of the genomic data identified two clearly discrete subpopulations, corresponding to the morphologically defined subspecies falcata and caerulea, with evidence of the hybrid nature of the subspecies hemicycla based on genome composition. Two distinct subpopulations exist within each subsp. caerulea and subsp. falcata. The distinction of caerulea was based on geographical distribution. The two falcata groups were separated based on ecogeography. The results show that taxonomic relationships based on morphology are reflected in the genetic marker data with some exceptions, and that clear distinctions among subspecies are evident at the diploid level. This research provides a baseline from which to systematically evaluate variability in tetraploid alfalfa and serves as a starting point for exploring diploid alfalfa for genetic and breeding experiments.





Similar content being viewed by others
References
Arnold ML, Hamrick JL, Bennett BD (1990) Allozyme variation in Louisiana irises: a test for introgression and hybrid speciation. Heredity 65:297–306
Barnes DK, Bingham ET, Murphy RP, Hunt OJ, Beard DF, Skrdla WH, Teuber LR (1977) Alfalfa germplasm in the United States: genetic vulnerability, use, improvement, and maintenance. USDA Tech Bull 1571
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brummer EC, Kochert G, Bouton JH (1991) RFLP variation in diploid and tetraploid alfalfa. Theor Appl Gen 83:89–96
Diwan N, Bouton JH, Kochert G, Cregan PB (2000) Mapping of simple sequence repeat (SSR) DNA markers in diploid and tetraploid alfalfa. Theor Appl Genet 101:165–172
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Flajoulot S, Ronfort J, Baudouin P, Barre P, Huguet T, Huyghe C, Julier B (2005) Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a breeding program, using SSR markers. Theor Appl Genet 111:1420–1429
Gross BL, Schwarzbach AE, Rieseberg LH (2003) Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae). Am J Bot 90:1708–1719
Havananda T, Brummer EC, Maureira-Butler IJ, Doyle JJ (2010) Relationships among diploid members of the Medicago sativa (Fabaceae) species complex based on chloroplast and mitochondrial DNA sequences. Syst Bot 35:140–150
Ivanov AI (1988) Alfalfa. Amerind Publishing, New Delhi
Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C (2003) Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol 3:9
Kidwell KK, Austin DF, Osborn TC (1994) RFLP evaluation of nine Medicago accessions representing the original germplasm sources for North American alfalfa cultivars. Crop Sci 34:230–236
Lesins K, Lesins I (1979) Genus Medicago (Leguminasae): A taxogenetic study. Kluwer, Dordrecht
Liu J (2002) POWERMARKER—a powerful software for marker data analysis. Raleigh, NC: North Carolina State University, Bioinformatics Research Center. http://www.powermarker.net
Ma X-F, Szmidt AE, Wang X-R (2006) Genetic structure and evolutionary history of a diploid hybrid pine Pinus densata inferred from the nucleotide variation at seven gene loci. Mol Biol Evol 23:807–816
Maureira IJ, Ortega F, Campos H, Osborn TC (2004) Population structure and combining ability of diverse Medicago sativa germplasms. Theor Appl Genet 109:775–782
McCoy TJ, Bingham ET (1988) Cytology and cytogenetics of alfalfa. In: Hanson AA, Barnes DK, Hill RR (eds) Alfalfa and alfalfa improvement. ASA-CSSA–SSSA, Madison
Mengoni A, Gori A, Bazzicalupo M (2000) Use of RAPD and microsatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa, Medicago sativa. Plant Breed 119:311–317
Michaud R, Lehman WF, Rumbaugh MD (1988) World distribution and historical development. In: Hanson AA, Barnes DK, Hill RR (eds) Alfalfa and alfalfa improvement. ASA-CSSA-SSSA, Madison
Muller MH, Prosperi JM, Santoni S, Ronfort J (2003) Inferences from mitochondrial DNA patterns on the domestication history of alfalfa (Medicago sativa). Mol Ecol 12:2187–2199
Muller MH, Poncet C, Prosperi JM, Santoni S, Ronfort J (2005) Domestication history in the Medicago sativa species complex: inferences from nuclear sequence polymorphism. Mol Ecol 15:1589–1602
Peakall R, Smouse PE (2001) GenAlEx V6: genetic analysis in excel population genetic software for teaching and research. Australian National University, Canberra. http://wwwanueduau/BoZo/GenAlE
Pritchard JK, Stevens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pupilli F, Businelli S, Paolocci F, Scotti C, Damiani F, Arcioni S (1996) Extent of RFLP variability in tetraploid populations of alfalfa, Medicago sativa. Plant Breed 115:106–112
Pupilli F, Labombarda P, Scotti C, Arcioni S (2000) RFLP analysis allows for the identification of alfalfa ecotypes. Plant Breed 119:271–276
Quiros CF, Bauchan GR (1988) The genus Medicago and the origin of the Medicago sativa complex. In: Hanson AA, Barnes DK, Hill RR (eds) Alfalfa and alfalfa improvement. ASA-CSSA-SSSA, Madison
Robins JG, Luth D, Campbell TA, Bauchan GR, He C, Viands DR, Hansen JL, Brummer EC (2007) Genetic mapping of biomass production in tetraploid alfalfa (Medicago sativa L). Crop Sci 47:1–10
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Segovia-Lerma A, Cantrell RG, Conway JM, Ray IM (2003) AFLP-based assessment of genetic diversity among nine alfalfa germplasms using bulk DNA templates. Genome 46:51–58
Sinskaya EN (1961) Flora of cultivated plants of the U.S.S.R. XIII perennial leguminous. National Science Foundation, Washington DC
Sledge MK, Ray IM, Jiang G (2005) An expressed sequence tag SSR map of tetraploid alfalfa (Medicago sativa L). Theor Appl Genet 111:980–992
Vandemark GJ, Ariss JJ, Bauchan GA, Larsen RC, Hughes TJ (2006) Estimating genetic relationships among historical sources of alfalfa germplasm and selected cultivars with sequence related amplified polymorphisms. Euphytica 152:9–16
Acknowledgments
We would like to thank Donald Wood, Jonathan Markham, and Frank Newsome for the technical assistance; the Turkish Government for funding MS; the USDA-DOE Plant Feedstock Genomics for Bioenergy award 2006-35300-17224 (to ECB and JJD) and National Science Foundation award DEB-0516673 (to JJD) for funding this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. -B. Veyrieras.
Rights and permissions
About this article
Cite this article
Şakiroğlu, M., Doyle, J.J. & Charles Brummer, E. Inferring population structure and genetic diversity of broad range of wild diploid alfalfa (Medicago sativa L.) accessions using SSR markers. Theor Appl Genet 121, 403–415 (2010). https://doi.org/10.1007/s00122-010-1319-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1319-4