Abstract
Sugar beet is hypothesized to have a narrowed genetic base due to its origin as White Silesian Beet and from numerous breeding selections and practices. High sugar quality, yield of recoverable sugar, cytoplasmic-male sterility system, monogermity, pests and disease resistance and bolting resistance constitute some of the adaptations that significantly influenced the existing genetic background of the crop. In this study we aimed to evaluate the extent of genetic diversity existing in wild beet representatives of Beta and Patellifolia and sugar beet cultivars, with a special focus on the complex Beta vulgaris. Another purpose was to determine the potential usefulness and conformity of selected molecular markers in different groups of materials in the context of rhizomania resistance. To reach these goals, molecular RAPD, ISSR techniques, literature-selected rhizomania resistance-segregating sequences as well as mitochondrial markers were used. The comparison of genetic diversity in wild and cultivated Beta forms shows that the population differentiation values and distance values are relatively high in cultivars. Moreover, the diversity component seemed to be compromised rather on the level of population (Hs) than in total (Ht) in cultivars. Our results shed a new light on the expected genetic bottlenecks existing in cultivars and revealed features specific for individual taxa (Patellifolia, Corollinae). Some degree of distinctiveness was suggested between genetic determinants of rhizomania resistance in modern cultivars in comparison with wild resistance sources. In addition, we document here an internal heterogeneity existing in selected wild/weedy accessions at the level of crucial sequences using high resolution melting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugar beet (Beta vulgaris L. ssp. vulgaris) is a crop with great economic importance, especially in temperate climate zones where it constitutes a valuable sucrose source. It contributes about one fifth to the global sugar production, which for the 2007–2011 period reached approximately 157 million metric tons (Koo and Taylor 2012). Crop rotation is necessary in beet production due to disease problems (Koo and Taylor 2012), which may also be effectively eliminated by resistance breeding approaches.
Breeding contributed above all to the increase in sugar yield. Some hybrids of fodder beets selected for relatively high sugar content, i.e. “White Silesian”, constituted the origin for all subsequently developed sugar beet varieties (Fischer 1989; Biancardi et al. 2010). At the same time other breeding practices and directions, for example cytoplasmic male sterility and genetic monogermity systems, used throughout the history of the modern crop’s development, are supposed to impinge significantly on its genetic structure and, as a consequence, to result in genetic bottlenecks, thus reducing available diversity and hampering in a way the progress in genetic combinations for new cultivars’ releases (Biancardi et al. 2010).
A considerably higher genetic diversity is expected to exist in wild relatives of sugar beet, serving thus as a reservoir of potentially useful traits and genes, such as those determining tolerance to biotic and abiotic stresses, yield parameters or other advantageous morphological and physiological features (Van Geyt et al. 1990; Stevanato et al. 2013). Hybridization between B. vulgaris and exotic germplasm belonging to Beta is possible, often providing fertile progeny, fully compatible at the chromosomal level. This phenomenon has been employed in sugar beet enhancement programs since the beginnings of the twentieth century, as growth in cultivated acreage and expanding distribution areas were inevitably related to increased pathogen pressure and disease spread (Panella and Lewellen 2007; Biancardi et al. 2010). From this time on, many directed and internationally coordinated initiatives have been undertaken by research and breeding communities, especially in France and the USA, aiming not only at the introgression of selected valuable traits into the sugar beet crop, but also at broadening of its genetic base. Such an integrated approach, although requiring further selection cycles, is at the same time expected to stimulate more rational and flexible management of genetic resources in the future (Frese et al. 2001). Additionally as a result of interfertile B. vulgaris coexisting in seed production areas, spontaneous outcrossings between wild and cultivated beets promote the origin of weed beets (Boudry et al. 1993). Despite its stochastic nature and basically unwanted effects in that particular case, the phenomenon itself may also have some positive implications, via increasing the variability level in the available gene pool.
One of the best examples for implementing desired characters successfully in wild sea beet (Beta vulgaris ssp. maritima) during breeding is rhizomania resistance genes. Early partially resistant sugar beet cultivars were developed based on Munerati’s pool of these accessions selected in the Po estuary for Cercospora leaf spot resistance (Van Geyt et al. 1990). Presently it is believed that all the resistance sources identified thus far, designated from Rz1 to Rz5, originate most probably from some wild sea beet ancestors (Biancardi et al. 2002), although their identity and independence from each other has not been fully confirmed yet.
In this study we aimed to evaluate the extent of genetic diversity existing in wild beet representatives belonging to the genera Beta and Patellifolia as compared to sugar beet cultivars with a special emphasis on the complex Beta vulgaris using molecular RAPD, ISSR techniques and literature-selected putative rhizomania resistance-segregating markers as well as mitochondrial markers.
Our results not only shed a new light on the expected genetic bottlenecks existing in cultivars and revealed some features specific for individual taxa (Patellifolia, Corollinae), but also suggest that some degree of distinctiveness may exist between genetic determinants of rhizomania resistance in modern cultivars when compared with wild resistance sources.
Materials and methods
Plant material
The materials selected for the study comprise representatives of five wild beet species, i.e. B. vulgaris ssp. maritima (4 accessions: B.m.01, B.m.27—resistant, B.m.40 and B.m.72), B. macrorhiza (B.macr.21), B. corolliflora (B.c.20), P. procumbens (P.p.25) and P. patellaris (P.pat.22), one weed beet accession (wbM), and five sugar beet cultivars with contrasting phenotypes for rhizomania resistance (B.v.14, B.v.15 and B.v.16—diploid, tolerant, B.v.13 and B.v.17—triploid, susceptible). Wild accessions were obtained from international genebanks, and a weed beet population was collected from a sugar beet field in Minikowo (Poland). Cultivated materials were obtained from commercial sugar beet breeding companies (Strube, KHBC Ltd.). Ten genotypes per each accession were individually analyzed, unless otherwise indicated. Further single individuals of wild B. vulgaris ssp. maritima/B. vulgaris ssp. vulgaris, representing the following accessions: B.m.28, B.m.29 (resistant), B.m.30, B.m.02, B.v.33 and B.m.31 (susceptible) were included as well for additional, more in-depth comparisons of the molecular marker efficiencies on an expanded group of materials and some for HRM (high resolution melting) standards also. In HRM analyses of cultivars, the standards were rhizomania resistance-segregating breeding materials kindly provided by KHBC Ltd. The list of accessions used in the study is presented in Table 1.
Genomic DNA isolation
DNA isolation from leaves was carried out as described previously (Davis et al. 1986). The concentration and integrity of obtained DNA samples were evaluated spectrophotometrically and electrophoretically.
PCR and electrophoresis
Primers for the study were RAPD (6), ISSR (6) and rhizomania resistance/tolerance-segregating sequences selected from the literature (18). PCR mixtures were as follows: 1 ng/µl DNA, 2.5 mM MgCl2 (Thermo Scientific), 0.2 mM dNTP (Thermo Scientific), 1 µM specific primers (Genomed), 0.56 u DreamTaq Polymerase (Thermo Scientific), 1× Taq Buffer with (NH4)2SO4 (Thermo Scientific) and PCR grade water up to 20 µl. Different reaction programmes were applied depending on the conditions previously described for a given primer sequence. Amplification products were separated on 1.5 % agarose (Prona Agarose, Basica Le) stained with ethidium bromide (Promega; 0.5 µg/ml) at 5 V/cm of gel. The size of the products was estimated using Gel Doc™ 2000 Gel Documentation System (Bio-Rad Laboratories Srl, Milan, Italy) equipped with Quantity One software, version 4.0.3, by comparison with the standard, i.e. GeneRuler™ 100 bp Plus DNA Ladder (Thermo Scientific). All the sequences of primers included in the study are presented in Table 2. Cytoplasmic diversity was examined using primers designed by Nishizawa et al. (2000) for mitochondrial minisatellites according to the protocol described previously by Fénart et al. (2008).
HRM analysis
Experiments were performed on CFX96™ Real-Time System (BIO-RAD) using AmpliQ 5× HOT EvaGreen® HRM Mix (Novazym). Primer sequences were designed using Primer BLAST (NCBI).
Statistical analyses
Dendrograms of genetic distance were constructed based on the genetic similarity coefficient according to Nei and Li (1979). Distance values were compiled using the unweighted pair-group method with arithmetic averages (UPGMA) in STATISTICA software (StatSoft, Inc.). All the clearly recognizable bands detected within a given accession (per 10 genotypes) were considered as potentially representative for this accession. Percentages of polymorphic loci in a given accession (for all cultivated and wild accessions in the study) were counted independently for all loci in each category of the primers (RAPD, ISSR and segregating sequences) and assayed by Mann–Whitney U statistics (GraphPad Prism, GraphPad Software, Inc.). Additionally, more in-depth analyses were performed for B. vulgars ssp. vulgaris as well as B. vulgaris ssp. maritima under study using POPGENE software, version 1.31 (F.C. Yeh, R.-c. Yang, University of Alberta and T. Boyle, Centre for International Forestry Research 1999), based on 289 loci obtained for 10 representative primers, applying available single—and grouped populations descriptive statistics.
Results
Overall diversity pattern of Beta and Patellifolia
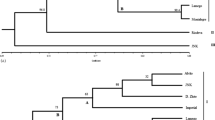
The median values of the mean percentages for polymorphic loci from Beta complex accessions under study were as follows: B. macrorhiza–73.74 %, B. corolliflora—66.67 %, B. vulgaris ssp. maritima—Greece (B.m.40), France (B.m.72), the Netherlands (B.m.01), Denmark (B.m.27)—66.3, 60.61, 57.8, 40.59 %, weed beet ‘Minikowo’ (wbM)—50.5 %, sugar beet cultivars—B.v.17, B.v.14, B.v.16, B.v.15, B.v.13—37, 32.81, 28.81, 27.74 and 26.14 %, respectively (Fig. 1). To better resolve the relationships among the investigated materials, a dendrogram of genetic distance was constructed (Fig. 2). Interestingly, based on this, linkage distances obtained for B. vulgaris ssp. maritima accessions of the different above-mentioned geographic origins did not seem to exceed the respective distance values observed within the cultivar group.
Genetic diversity for the accessions under study expressed as the percentage of polymorphic loci (PCR products for all the primers included). a—all the accessions examined: 1 P. patellaris, 2 P. procumbens, 3 B. vulgaris ssp. maritima (B.m.27), 4 weed beet ‘Minikowo’ (wbM), 5 B. vulgaris ssp. maritima (B.m.01), 6 B. vulgaris ssp. maritima (B.m.72), 7 B. vulgaris ssp. maritima (B.m.40), 8 B. corolliflora, 9 B. macrorhiza, 10–14 sugar beet cultivars: 10 B.v.13, 11 B.v.15, 12 B.v.16, 13 B.v.14, 14 B.v.17. b–h comparisons in selected groups of accessions; 1 wild/weed beet group [b—all the wild accessions under study, c—B. macrorhiza, d—B. corolliflora, e—B. vulgaris ssp. maritima (B.m.27, B.m.01, B.m.72, B.m.40), f—weed beet ‘Minikowo’ (wbM), g—P. procumbens, h—P. patellaris], 2 cultivar group. Statistically significant differences were recognized in all cases from b to h except for g (Mann–Whitney U, P < 0.05). Columns—median percentage, bars—interquartile range
Restricted genetic diversity levels, expressed as the polymorphic loci percentage and comparable to cultivated beet representatives, were encountered in Patellifolia with the median values of 36.67 and 17.65 % for P. procumbens and P. patellaris, respectively (Fig. 1). In addition, accessions of this genus formed a visibly distinct cluster in the dendrogram of genetic distance (Fig. 2). In contrast, a low genetic distance obtained between the B. corolliflora and B. macrorhiza accessions was accompanied by the highest frequency of polymorphic loci per accession. Genetic distance between these two forms falls within a slightly lower range of values than that obtained for the most similar sugar beet cultivars (Fig. 2).
In order to perform a more in-depth comparison of the differences found in the investigated populations of Beta complex, we applied also single-population and grouped populations descriptive statistics using POPGENE version 1.31, based on 10 selected primers for B. vulgaris ssp. maritima, wbM and cultivars (Tables 3, 4). According to Table 3, in case of B. vulgaris ssp. maritima accessions and weed beets the same tendency of genetic diversity was retained as for the median percentages of polymorphic loci obtained for all the primers used. Slight differences in the ordering of accessions was found for the cultivar group, which might be accounted for by their relatively comparable median values.
Beta vulgaris complex
Mitochondrial diversity pattern
In our study we identified seven different mitochondrial haplotypes in Beta vulgaris group, out of which six were represented by B. vulgaris ssp. maritima accessions (Table 5). We confirmed here the presence of seven mitotypes bearing resemblance to those described by Fénart et al. (2008), four of which showed identical banding patterns, i.e. C (G*), D (A/I*), F (?*) and Owen (Owen CMS*), as well as three with similar banding patterns, i.e. A (?*?), B (H/N*?) and E (F*?). The haplotypes in B. vulgaris ssp. maritima were defined upon recognition of 5 alleles for TR1, 2 for TR2, 1 for TR3 and 2 for TR4. According to TR1-4 minisatellite pattern, the occurrence of the Owen-type cytoplasm was documented for the weed beet population as well as for all the cultivars under study.
Sugar beet cultivars
Here we found that differences in the percentage of polymorphic loci were statistically significant (Mann–Whitney U P < 0.05) between sugar beet cultivars (median: 32.14 %) and the wild/weedy group (median: 57.32 %) (Fig. 1b–f), with the least pronounced difference among Beta complex in case of the cultivars-weed beet comparison (Fig. 1f). These results were further confirmed and extended by more detailed analyses suggesting that a significant component of the difference between Group II represented by cultivars and Group I represented by wild/weed beets lies within the mean diversity within population (Hs) as compared with total genetic diversity in the pooled populations (Ht) (Table 4). Cultivated materials featured a higher population differentiation measure (Gst) than did wild/weedy populations.
Based on the dendrogram of genetic distance (Fig. 2), the materials obtained from different suppliers (B.v.13 and B.v.17 vs. B.v.14, B.v.15 and B.v.16) display relatively close location to one another, so that one of them, i.e. B.v.16, seemed to cluster with the former group rather than with the latter.
The differences in banding patterns between individual genotypes within a single cultivar were encountered, although with a frequency lower than in their wild counterparts. What is more, we obtained 12 original products that allow to distinguish individual cultivars among the group tested (data not shown), being indicative for a particular cultivar and thus useful for genotype identification in this group.
Weed beets
Molecular analyses performed in this study revealed some unique features of the weed beet population collected from a sugar beet field near Minikowo (Poland). As compared with cultivars, the material was characterized by relatively high genetic diversity measures (Fig. 1—median percentage of polymorphic loci: 50.5 %; Table 3-h = 0.0864, I = 0.1297, polymorphic loci: 73 %) and a closer resemblance to wild B. vulgaris ssp. maritima populations as far as molecular profiles are concerned, which was evident from its clustreing in the dendrogram of genetic distance (Fig. 2). At the same time cytoplasmic uniformity was observed (Table 5).
Another accession included in this study that bears characteristics of a weedy population is B.m.40, according to IDBB information. However, due to its primary taxonomic classification, geographic origin (based on a genebank passport data), mitochondrial DNA pattern and dendrogram location based on molecular profiles obtained in our study (Fig. 2), it was described below.
Beta vulgaris ssp. maritima
In this study we obtained a rather wide range of values as regards the overall genetic diversity or allelic richness represented by B. vulgaris ssp. maritima accessions differing in their origin. The borderline values in this group were as follows: from 0.1105 to 0.417 for Nei’s gene diversity, from 0.1648 to 0.0616 for Shannon’s index and from 89 to 33 % for polymorphic loci, see Table 3.
Interestingly, the highest degree of gene diversity and the highest percentages of polymorphic loci were found in B.m.40 and B.m.72 accessions (Fig. 1; Table 3) that displayed the highest number of mitochondrial haplotypes as well, i.e. three different per ten genotypes for each of these accessions, whereas in other B. vulgaris ssp. maritima accessions we recorded only one haplotype per ten genotypes of each accession (Table 5).
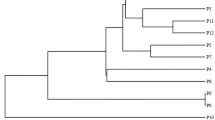
Moreover, using a set of selected rhizomania resistance-segregating primers in the extended group of B. vulgaris accessions, we constructed another dendrogram and identified the particularly low genetic distance (0.2) for some B. vulgaris ssp. maritima accessions that were known as the sources of rhizomania resistance (B.m.28, B.m.29) and thus may in fact be closely related (Fig. 3). Interestingly, another highly resistant accession (B.m.27) clustered with the group of other phenotypically undefined accessions, including B.m.01, wbM and B.m.72.
Dendrogram of genetic distance for the extended group of wild B. vulgaris ssp. maritima/B. vulgaris ssp. vulgaris/weed beet genotypes representing different accessions, constructed using UPGMA cluster analysis (unweighted pair-group method using arithmetic averages), based on the data obtained for six selected phenotype-segregating primers
Rhizomania resistance-segregating markers
Using rhizomania resistance-segregating sequences previously described (Table 2) for the cultivars of contrasting phenotypes we were not able to confirm the presence of the same size fragments that were previously recognized as rhizomania resistance-segregating. On the contrary, such sequences were found by us in wild accessions. For example, the presence of one of resistance gene analogues that was previously shown to be common and universal for different resistance sources (Lein et al. 2006) was not documented here for the cultivar group, but was typical for resistant B. vulgaris ssp. maritima accessions (Fig. 4). Instead, in the cultivar group, other bands appeared in the number of 15, manifesting a high rate of conformity with the phenotype.
PCR analysis for the presence of the resistance gene analogue cZR-3 in DNA of the extended group of wild B. vulgaris ssp. maritima/B. vulgaris ssp. vulgaris/weedy genotypes. L—ladder, C—negative control, 1 B.m.02, 2 B.m.28, 3 B.m.29, 4 B.m.30, 5 B.m.31, 6 B.v.33, 7 B.m.01, 8 weed beet ‘Minikowo’ (wbM), 9 B.m.27, 10 B.m.40, 11 B.m.72. The arrow points to the expected product, visible in lines: 2 (B.m.28), 3 (B.m.29), 7 (B.m.01) and 9 (B.m.27)
We performed also high resolution melting analysis (HRM) after PCR for one of the products in order to identify potential Rz2-accompanying polymorphisms described earlier (Thiel et al. 2012). As a result we found that one of the resistant cultivars repeatedly clustered with the susceptible-like profile as far as this sequence is concerned, displaying a heterozygote-like HRM profile (Fig. 5a). General orientation of the differences was inverted in cultivars and breeding materials as compared to the wild beet group despite identical melting curves (Figs. 5a vs. 5b–f, 6), which implies greater sequential heterogeneity in the wild counterparts and/or the involvement of other/additional resistance-determining factors. Our investigation of ten genotypes per each wild B. vulgaris ssp. maritima and weedy accession showed that interesting profiles may be distinguished in these materials, some of which displayed conformity with the profiles previously reported by us for highly resistant accessions, e.g. in B.m.72 (Fig. 5b; Litwiniec et al. 2014). Some of the studied wild/weed accessions feature an internal diversity on the level of the fragment examined by HRM, which was original and repeatable for each genotype under study. As this phenomenon was especially evident for the accession B.m.01 and the weed beet ‘Minikowo’, which segregated in its profiles into putative resistant-like or susceptible-like groups (Fig. 5d, e), we would like to suggest their potential usefulness for further experiments and underline that not only wild, but also weed beets may constitute a potential source of new useful traits.
Representative high resolution melting (HRM) profiles obtained for the investigated product (an Rz2-accompanying polymorphism) on the templates under study. a—cultivars and breeding standards (the arrow points to a putative heterozygous cultivar, B.v.15, which along with B.v.13 and B.v.17 group together—blue profiles, as opposed to B.v.14 and B.v.16—green profiles), a’—breeding standards (RR—green, Rr—blue and rr—red). Heterozygotes and/or homozygotes are distinguished based on their HRM profile shape and RFU (relative fluorescence units) difference values as compared to the respective standards. b–f wild beets B. vulgaris ssp. maritima/weed beets: b B.m.72, c B.m.27, d B.m.01, e weed beet ‘Minikowo’ (wbM), f B.m.40; and b’–f’ their respective standards (green —susceptible, red —resistant/tolerant). Visible segregation in the following accessions: d B.m.01, e weed beet ‘Minikowo’ (wbM). The axes stand for Y (vertical): difference RFU, X (horizontal): temperature (Celsius degrees). (Color figure online)
Representative melting curve profiles obtained for the investigated product (an Rz2-accompanying polymorphism) on templates of a cultivars and breeding standards, b wild beets B. vulgaris ssp. maritima and breeding standards. The axes stand for Y (vertical): -d(RFU)/dT, X (horizontal): temperature (Celsius degrees). RFU relative fluorescence units
Discussion
Overall diversity pattern of Beta and Patellifolia
Molecular DNA markers are widely used to evaluate genetic relationships between plant taxa as well as their linkage to particular phenotypic features for marker assisted-selection (Winter and Kahl 1995). Besides, it is believed that comparison of different marker types, i.e. arbitrary and repetitive as well as these related to specific traits, gives a more detailed insight into the direction of selection and composition of natural populations (Richards et al. 2004), which in our study was also extended to commercially-produced sugar beet representatives. Due to the risks associated with disease spread in highly uniform populations, our aim was to reveal the genetic structure represented by present cultivar germplasm in relation to their wild counterparts using neutral and specific rhizomania resistance-related molecular markers. This is therefore expected not only to broaden our understanding of phylogenetic relationships and general genetic diversity within the investigated taxa, but also to provide information concerning the occurrence of potentially universal and/or accession-unique rhizomania resistance-segregating sequences, the linkage of which may have been broken over the course of evolution and domestication. Thus, we believe that comparison of wild, weed and cultivated beets using different molecular markers’ categories may give us a more comprehensive insight into the scale and direction of genetic bottlenecks inevitably accompanying progress in breeding this crop.
Previously, neutral molecular markers were also broadly employed for characterization of Beta and Patellifolia genetic resources (Reamon- Büttner et al. 1996; Kubis et al. 1997; Smulders et al. 2010). These techniques allowed precise assessment of the genetic diversity existing in cultivated and wild beet relatives and suggested that a considerable pool of alleles was lost during the domestication process as compared to wild sea beets which probably bear more resemblance to sugar beet ancestors, eventually leaving as low as one third to one fourth of the available natural variation (Richards et al. 2004; Fénart et al. 2008; Saccomani et al. 2009; Biancardi et al. 2010).
This assumption is reflected by our results to some extent, as we documented here a statistically significantly lower percentage of polymorphic loci in cultivated materials when compared with wild beet species. This was especially evident with reference to the representatives of the section Corollinae and B. vulgaris ssp. maritima, i.e. other species belonging to the present genus Beta. The median values of the mean percentages of polymorphic loci as well as linkage distances in the dendrogram obtained by us suggest that sugar beet cultivars still represent a relatively well genetically differentiated group, but with a lower ratio of an internal within-cultivar heterogeneity. This supposition was further confirmed by grouped populations descriptive statistics using POPGENE, as in the comparison of wild and weed beet groups (Group 1) to cultivars (Group 2), we found that although gene diversities either among subpopulations (Hs) or of the total population (Ht) were higher in the wild-weed group, the population differentiation values (Gst) were even compromised in relation to the cultivars. It might however have been expected, taking into account the sugar beet breeding system and especially the diversity delivered by pollinators.
Limited genetic diversity, as well as separate clustering in the dendrogram of genetic distance in case of Patellifolia in this study may indicate their early separation event from the putative common ancestry of the sections Beta and Corollinae as also previously reported (Kubis et al. 1997; Kadereit et al. 2006). Being restricted in their distribution pattern to the regions of Canary Islands, Madeira and Southern Iberian coast, Patellifolia display actually a closer relationship with the representatives of other genera grouping to Hablitzieae, therefore their classification into the Beta-independent taxonomic unit has been suggested (Scott et al. 1977; Jung et al. 1993; Kubis et al. 1997; Kadereit et al. 2006). The lowest frequency of polymorphisms found for P. patellaris may be explained by the fact that this is a self-fertile tetraploid form.
A low genetic distance obtained by us between Corollinae accessions is also noteworthy and may reflect the fact that this section constitutes actually a biogeographical complex of intermediate forms that can often cross easily with one another. Therefore, the high frequency of polymorphic loci per accession is in apparent contradiction with the low distance values in this case only. Although B. macrorhiza and B. corolliflora are generally regarded to represent separate forms/species, it seems at the same time that the borders between individuals belonging to this section may in fact be less clearly delineated and much more difficult to recognize than for any other exotic germplasm combination tested in our study. As proved by molecular methods, such a close relationship between these species was reported earlier (Jung et al. 1993). Some accessions belonging to these species cluster with each other and as a consequence may be easily misclassified (Reamon-Büttner et al. 1996).
Beta vulgaris complex
Mitochondrial diversity pattern
A considerable degree of homogeneity found in cultivated beets may be on account of the introduction and ubiquitous exploitation of common sources of advantageous traits in the history of breeding, i.e. bolting or disease resistance, monogermity and the Owen’s source of cytoplasmic male sterility. The literature reports nevertheless on the possibility of identification of various alternative similar systems in wild beets and in sugar beet (Dalke and Szota 1986; Van Geyt et al. 1990). For example, as far as mitochondrial minisatellite haplotypes are concerned, the study by Fénart et al. (2008) reflects a sharp contrast between cultivars/weed beets, representing predominantly the Owen type of cytoplasm, and wild ruderal/sea beets for which ten different mitotypes were recognized. Therefore, maternal cultivated origin of the vast majority of weed beets investigated was documented there, except for approximately 11 % of the individuals tested (Fénart et al. 2008). We confirmed here greater mitochondrial DNA heterogeneity in B. vulgaris ssp. maritima group as opposed to the uniformity found in the cultivated cytoplasm and/or respective descendants of this, i.e. weed beets.
Sugar beet cultivars
With regard to nuclear components, allelic richness was shown to be the lowest thus far in the cultivated beet group. At the same time the highest population fixation indices were obtained either within wild sea beet populations or between this group and the other forms examined (Desplanque et al. 1999; Fénart et al. 2008). Moreover, different neighbour-joining trees indicated at the clear distinctiveness between the last mentioned group and the other clusters including wild ruderal and/or weed beets, thus implying the earliest evolutionary divergence at this point of the complex as well as a relatively higher probability of gene flow between wild ruderal populations, weed beets and cultivars with a special emphasis on the two latter (Desplanque et al. 1999; Fénart et al. 2008; Saccomani et al. 2009).
In our study we confirmed that the percentage of polymorphic loci was significantly lower for sugar beet cultivars than for wild or weed beet populations, which was further supported by the Nei’s gene diversity (1973) measure or Shannon’s information index (Lewontin 1972; Nei 1973, 1987). Nevertheless, some differences in banding patterns between individual genotypes belonging to the same cultivar were observed by us, and may be found more often for materials representing different breeding lines, which was documented earlier for instance using satellite DNA repeats (Kubis et al. 1997). It seems that the differences in banding patterns observed within the same cultivar result most probably from segregation for a trait that was not among the selection criteria for the development of this particular cultivar. Previously McGrath et al. (1999) performed a more in-depth analysis of genetic diversity in selected US sugar beet releases, lines and breeding materials in comparison with some wild B. vulgaris ssp. maritima accessions. They found that although total heterozygosity of the sugar beet accessions had not changed significantly over the period of several decades, breeding practices had a substantial impact on diversity reduction within releases. The most important factors contributing to this reduction included self-fertility, monogermity and other characters essential for the production of hybrid seeds, e.g. alleles conditioning O-type lines maintenance. As diversity was rather structured or sub-divided within accessions, it has been hypothesized that breeding progress is still possible, but depends on respective selection of parental components from different accessions and supplementation of the genetic pool with wild-originating advantageous alleles. The study showed also separate grouping of wild materials based on the dendrogram of similarity and relatively high rate of heterozygosity within populations of wild beets. Nevertheless their total heterozygosity was underestimated compared to sugar beet accessions due to the scoring approach undertaken (McGrath et al. 1999). Similar to McGrath et al. (1999) we found also that the difference between cultivars and wild/weed beets was far more pronounced at the level of the mean diversity within each population, than for the total genetic diversity in the pooled populations. Moreover, in this study we report the relatively high population differentiation measure in the cultivar group. In a broader phylogenetic background, our results show that the level of genetic variation expressed as the percentage of polymorphic loci in sugar beet cultivars may be as low as the case of Patellifolia, which is currently ascribed to the separate genus and the representatives of which are believed to approach their final evolutionary state (Frese 1989 from Jung et al. 1993). Besides, we confirm here that a considerable part of biodiversity may still be exploited from the sections Beta and especially Corollinae with the latter constituting most probably the driving force of ongoing speciation, as it was previously reported (Kubis et al. 1997). These results are also in agreement with the observations made by Jung et al. (1993), according to which sugar beet cultivars manifest low values of genetic diversity and a close relationship to B. vulgaris ssp. maritima, but rather low genetic similarity to other wild species (Jung et al. 1993). Relatively comparable values of genetic distance were obtained by Mita et al. (1991) for sugar beet cultivars and selected wild accessions, nevertheless the lower number of accessions was included in that study (Mita et al. 1991).
In agreement with the lower internal within-cultivar heterogeneity in the studied subpopulations and in total, we found also some sequences that were original and non-polymorphic, hence potentially indicative of a particular cultivar, which therefore may be employed for fingerprinting purposes. The uttermost importance of this application for variety registration was previously raised (Jung et al. 1993; Smulders et al. 2010). We confirmed also a general tendency observed by Smulders et al. (2010), according to which diploid cultivars display greater genetic distances and higher levels of genetic differentiation than triploids due to a higher probability of allelic repetitions in the latter (Smulders et al. 2010). They underlined that another significant component of differentiation may be a breeding pool that constitutes the basis for a given material. In agreement with this, we found some distance differences in the dendrogram for the materials obtained from different suppliers, although they do not seem to form clearly distinct clusters. This may be explained by partial overlaps existing at present in different breeding programs, caused for example by limited number of maternal lines available. As documented by Fénart et al. (2008), 13 cultivars derived from different seed producing companies featured rather low levels of genetic differentiation (Fénart et al. 2008). In our opinion however the presumption of Smulders et al. (2010) suggesting a putative lack of genetic bottlenecks appearing as a consequence of breeding from wild ancestors, should rather be viewed in light of the outstanding efficiencies of microsatellites developed by them for cultivated materials, as wild counterparts were not included for comparisons in their study. Though, the statement that the heterogeneity level maintained in the sugar beet materials is sufficient and may be accounted to the system of hybrid varieties seems fully justified, because the use of numerous parental lines contributes to different possible gene combinations in their descending pool. Another study that suggested comparable levels of genetic variation between sugar beet lines and wild relatives was based mostly on single copy DNA sequences, which were employed for the analysis of individual plants per accession (Hjerdin et al. 1994). In this study we evaluated the occurrence of multi-copied fragments across ten independent individuals per accession, which allowed us also to take a closer look at the rate of internal differentiation and allele richness. A proof for the usefulness of the molecular markers applied by us may be the highest percentage of polymorphic bands obtained for B. macrorhiza and B. corolliflora, displaying at the same time the lowest genetic distance values among analyzed accessions. The relatively low value of genetic distance obtained in this study between wild sea beets/weedy beets and cultivated forms reflect perhaps recent domestication history of the crop from its wild ancestors and continuous exchanges of the genetic material within the beet complex.
Weed beets
Another important subject in terms of genetic diversity of different evolving Beta system forms is the evaluation of the wild ruderal and weed beet populations in the context of their origin and the events of crop-to-wild or wild-to-crop gene flows which may occur in the vicinity of sugar beet seed production areas. Such introgressions may be the source of novel traits for breeding and result in increased genetic diversity on one hand. On the other, they also influence the wild beet genetic pool on the long run, thus creating potential risks for a transgene escape and/or gradual depletion in allelic richness as a consequence of swamping with more invasive forms (Bartsch et al. 1999; Viard et al. 2004; Arnaud et al. 2009). Arnaud et al. (2009) found for example that there was a dilution of crop nuclear genes in contrast to cytoplasmic pool in inland wild populations of the former French seed production areas, thus suggesting past seed escapes. Since weed beets result from accidental hybridizations between wild and cultivated forms, they represent a genetically differentiated group as compared to wild beets and may be expected to display an intermediate position in the dendrogram of genetic distance. What is more, they often manifest a relatively high allelic richness, thus more closely resembling their wild counterparts than cultivated beets in this particular context (Desplanque et al. 1999; Viard et al. 2004; Fénart et al. 2008), which was also evident in our study for the wbM population. Besides, genetic isolation and relatively high heterozygote deficiencies have been reported in many weed beet populations, suggesting their independent origins. Other important features include self-fertility inheritance and substructuring within a population (Viard et al. 2002, 2004; Fénart et al. 2008). It is believed that these complexes usually consist of a mixture of spatially and temporally isolated genotypes, potentially resulting from multiple recurrent hybridization events between diverse sources, including a long-lived seed bank in the field, but also cross-pollinating or selfing in a density-dependent pattern (Fénart et al. 2007). In our study we confirmed the intermediate position of a weed beet population under study, i.e. the accession mentioned clustered in the dendrogram of genetic distance with the group of wild B. vulgaris ssp. maritima accessions and displayed comparable coefficients of genetic diversity. Its location was nevertheless the most distant within this group and as a result also visibly closer to the cluster of cultivars than that for any other wild/weedy beet examined. Several mitotypes can be potentially found within weedy populations, most probably derived from wild or ancient cultivated materials (Viard et al. 2004; Fénart et al. 2008). However, the most common for cultivated beets Owen CMS mitotype that was identified by us, suggests a cultivated maternal origin of the weedy accession under study. We analyzed one population of weed beets found in the sugar beet field near Minikowo (Poland), thus most probably originating from pollinations of cultivated beets in the seed production areas with wild pollen donors and subsequently accidentally introduced to Poland with seed lots. Therefore we expect that the materials may be comparable to those previously shown as genetically differentiated from wild ruderal and sea populations (Desplanque et al. 1999; Fénart et al. 2008). On the other hand, it has been explained previously that some of weed beets clustering into the group of cultivars may in fact represent bolters with low vernalization requirements (Fénart et al. 2008). Although it could not be ruled out in our study as well, to avoid this we classified the weed beet population based on several different criteria, i.e. atypical morphology of the root, anthocyanin pigmentation, as well as out-row location, raising the possibility that this material represents some genetic components of a local long-lasting seed bank rather than constituting solely a seasonal population introduced to Poland with a lot of seeds, and thus putatively displays higher genetic diversity and closer resemblance to wild counterparts than cultivars, which was actually confirmed. We believe that it is worth to extend this studies, especially in Poland, where diversity of local weedy beet populations is poorly addressed and for the reasons of their interesting molecular profiles that were mentioned before. More detailed analyses devoted to sugar beet cultivars in comparison with in-row and out-row weedy beets in France suggested that as far as 25–29 % of in-row weedy beets may in fact constitute bolting cultivars and that genetic diversity in this complex increased gradually from the cultivar to out-row weeds’ component, based on biparentally inherited markers and in sharp contrast to the homogeneity of cytoplasmic pool of genes (Viard et al. 2002). The above-mentioned study indicates that a significant genetic differentiation may be observed even between in-row and out-row beets as well.
Beta vulgaris ssp. maritima
At the same time, however, genetic distinctiveness between inland weed and inland wild populations as well as cultivated and ruderal forms may be less pronounced in relation to the respective values obtained for inland wild and wild coastal populations, especially the Atlantic Coast populations (Desplanque et al. 1999; Fénart et al. 2008; Arnaud et al. 2009; Saccomani et al. 2009). Wild coastal populations are mostly differentiated depending on the location and influenced by variable ecological habitats (Desplanque et al. 1999; Saccomani et al. 2009). Their genetic pool may also be regionally exchanged with cultivated materials or ruderal beets, as it was suggested for example for the West Adriatic and Mediterranean accessions of sea beet, which may in fact constitute the origin of inland populations. Similar events are reported for other regions where sugar beet fields are in close proximity to the wild coastal beets (Bartsch et al. 1999; Desplanque et al. 1999; Viard et al. 2004; Fénart et al. 2008; Stevanato et al. 2013). As far as B. vulgaris ssp. maritima populations are concerned, especially high efficiency in discrimination of spatial differentiation between different populations of B. vulgaris ssp. maritima was found earlier for mitochondrial minisatellite loci (Fievet et al. 2007). In our study we confirmed the relatively high diversity of mitochondrial DNA in B. vulgaris ssp. maritima accessions, as six out of seven haplotypes were identified in this group, without the evidence of Owen CMS cytoplasm. Previous studies showed that the Owen CMS cytoplasm may exceptionally be encountered in wild sea beet populations and reflect seed-mediated transfer of cultivated genes (Viard et al. 2004). In contrast to our observations regarding the overall genetic diversity or allelic richness, displaying the rather wide range of values for the accessions of B. vulgaris ssp. maritima differing in their origin, these parameters should supposedly be quite similar for closely located populations, in which there is a high probability of gene flow. This was actually the case for French coastal and insular populations, for which surprisingly no clear geographical clustering in the dendrogram was documented, unless nuclear microsatellite data were also contained apart from cytoplasmic minisatellites. Using these markers more precise genetic boundaries were delineated, which turned out to be consistent with marine currents, and the continental origin of insular populations was suggested (Fievet et al. 2007). For the accessions of B. vulgaris ssp. maritima that are collected from distant geographic locations, a broad range of variability is usually encountered (Jung et al. 1993), as they most probably represent different centers of crop diversity. This tendency was also expected, but only partially reflected in our study, as the linkage distances observed between different B. vulgaris ssp. maritima accessions were in fact no significantly higher than those of different sugar beet cultivars. However, at the same time we recognized that a tendency concerning the occurrence of polymorphic products is confirmed by geographic location of the particular accession. For example, the highest genetic diversity expressed as the percentage of polymorphic bands was found for B. vulgaris ssp. maritima derived from Greece (B.m.40) as compared to other accessions, which was also confirmed by its morphological characteristics (data not shown). This observation supports the theory claiming that an exceptionally high heterogeneity may be encountered in the Mediterranean area, especially in a close proximity to the overlapping borders of distribution of the section Beta and Corollinae, which are regarded to constitute a center of speciation and differentiation among the existing genetic pools. However, it could not be excluded that in case of B.m.40 accession, this high diversity effect may also be enhanced by a weedy origin of this particular accession to some extent (according to IDBB).
Rhizomania resistance-segregating markers
In case of rhizomania resistance there are many reports concerning identification of potentially useful molecular markers that turned out to segregate with the feature in different segregating populations and, as such, they may be potentially efficient tools for breeders (Barzen et al. 1997; Giorio et al. 1997; Scholten et al. 1997; Gidner et al. 2005; Lein et al. 2006; Nouhi et al. 2008; Amiri et al. 2009; Feghhi et al. 2012). However, functional significance for most of them is poorly understood yet, and all the more so because most of them may not necessarily be located within a single major gene. It could not be ruled out that different additional sequences may be typical for a particular population only and, as such, usually do not feature the universal character across all the resistant materials. What is more, a hypothesis of different resistance sources have been raised, but at the same time it is not fully consistent yet, at least in some cases, whether they are conditioned by independent loci or by genes that represent in fact an allelic series. For example, over the course of the experimental crosses between Rz1- and Rz2-carrying materials (Holly-1-4 and WB42) it was determined that the distance between these genes may be as high as 20 to 35 cM (Scholten et al. 1999; Amiri et al. 2003), however the distinction between these genes and closely located Rz3 identified in WB41 may not be so unequivocal (Gidner et al. 2005). On the other hand, the situation seems to be more complex, as there are reports suggesting the presence of further genes, such as Rz4 and Rz5 (Grimmer et al. 2007, 2008). Here we propose the evaluation of efficiency of a panel of molecular markers for the materials of different origins, i.e. wild, weed beets as well as cultivars with contrasting rhizomania-resistance phenotypes.
We verified the usefulness and uniformity of selected rhizomania resistance-segregating markers described in the literature across a broad background of beet materials. As far as these sequences are concerned, it is regarded that different commercially used proprietary sequences are applied by various breeding companies (Biancardi et al. 2010). In our study, we aimed to evaluate the efficiency of the available sequences in the context of potential usefulness for marker-assisted selection and discrimination of characteristics that would be indicative of a given resistance source. Interestingly, we proved here a distinctiveness existing between wild crop relatives on one hand and modern sugar beet cultivars on the other, thus implying the contribution of some unique/original sequences in each of these groups and raising the possibility of partly independent bases of the resistance represented by these materials.
Our present results most probably reflect the fact that to some extent different contributing genes and mechanisms may have been employed in sugar beet cultivars in order to trigger the resistance response as compared to the wild group, which may for example be caused by incompletely inherited pool of resistance determinants. It seems consistent with our previous observations as well (Litwiniec et al. 2014), according to which there is rather a huge discrepancy of the resistance rates between wild sources of resistance and tolerant cultivars. On the other hand, bearing in mind the polymorphisms found in some universal sequences of wild sea beet (B. vulgaris ssp. maritima), cultivars and traditional breeding materials, these observations may be in accord with the above cited hypothesis of rhizomania resistance origin. However, they may also suggest that in cultivars some secondary resistance determining mechanisms are more common, like those Polymyxa betae-targeting or programmed cell death-inducing, whereas still other specific or at least much more efficient contributors probably exist/prevail in resistant wild beet materials, resulting in their outstanding phenotypic manifestations or near-immunity (Litwiniec et al. 2014). One explanation for this may be that a complex defense pathway that is functional in the genetic background of a wild beet may not be easily transferred and maintained in the cultivated forms due to backcrosses and repeated selection cycles that are definitely required to eliminate the load of negative wild beet traits. Therefore, further careful parallel comparisons of genotypic and phenotypic characters in wild sources of resistance, but also in their respective breeding descendants, is absolutely essential for identification and understanding of the function and limitations of the factors that are crucial for resistance development in different genetic backgrounds. Engineering of the most efficient, complete metabolic or defense pathways would be the challenge of the future (Krichevsky et al. 2012) and requires thorough basic research in order to identify their indispensable components.
Conclusion
The knowledge on available biodiversity including classification and identification of the existing resistance sources will constitute a prerequisite for rational management of genetic resources. We believe that the initial step on that way may be not only selection, evaluation and maintenance of valuable genotypes, but above all the distinction between general and specific resistance bases in the materials of different origins. At the same time, our results suggest that the modern sugar beet cultivars may be partially narrowed and/or altered in terms of their resistance determinants as compared to wild beet materials, which however seems an inevitable tendency, taking into account the above-mentioned wild germplasm-related obstacles on one hand and the ever-increasing demands of market competitiveness on the other. Therefore, different initiatives have been undertaken thus far to broaden the genetic base of Beta (Frese et al. 2001). Apart from the impact of breeding practices, another important point contributing to discrepancies between wild and cultivated germplasms is that diverse selection pressures of native wild beet habitats favour the emergence and preferential maintenance of new advantageous alleles/polymorphisms, for instance in response to resistance-breaking viral pathotypes.
References
Amiri R, Moghaddam M, Mesbah M, Sadeghian SY, Ghannadha MR, Izadpanah K (2003) The inheritance of resistance to beet necrotic yellow vein virus (BNYVV) in B. vulgaris subsp. maritima, accession WB42: statistical comparisons with Holly-1-4. Euphytica 132:363–373
Amiri R, Mesbah M, Moghaddam M, Bihamta MR, Mohammadi SA, Norouzi P (2009) A new RAPD marker for beet necrotic yellow vein virus resistance gene in Beta vulgaris. Biol Plant 53:112–119
Arnaud J-F, Fénart S, Godé C, Deledicque S, Touzet P, Cuguen J (2009) Fine-scale geographical structure of genetic diversity in inland wild beet populations. Mol Ecol 18:3201–3215
Bartsch D, Lehnen M, Clegg J, Pohl-Orf M, Schuphan I, Ellstrand NC (1999) Impact of gene flow from cultivated beet on genetic diversity of wild sea beet populations. Mol Ecol 8:1733–1741
Barzen E, Stahl R, Fuchs E, Borchardt DC, Salamini F (1997) Development of coupling-repulsion-phase SCAR markers diagnostic for the sugar beet Rr1 allele conferring resistance to rhizomania. Mol Breed 3:231–238
Biancardi E, Lewellen RT, De Biaggi M, Erichsen AW, Stevanato P (2002) The origin of rhizomania resistance in sugar beet. Euphytica 127:383–397
Biancardi E, McGrath JM, Panella LW, Lewellen RT, Stevanato P (2010) Sugar beet. In: Bradshaw JE (ed) Root and tuber crops, handbook of plant breeding 7. Springer Science+Business Media, New York, pp 173–219
Boudry P, Mörchen M, Saumitou-Laprade P, Vernet Ph, Van Dijk H (1993) The origin and evolution of weed beets: consequences for the breeding and release of herbicide-resistant transgenic sugar beets. Theor Appl Genet 87:471–478
Dalke L, Szota M (1986) A search for new sources of male sterility for breeding hybrid sugar beet varieties. Genet Polonica 27:81–88
Davis LG, Dibner MD, Battey JF (1986) Preparation of DNA from eukaryotic cells. Basic methods in molecular biology. Elsevier, New York, pp 42–50
Desplanque B, Boudry P, Broomberg K, Saumitou-Laprade P, Cuguen J, Van Dijk H (1999) Genetic diversity and gene flow between wild, cultivated and weedy forms of Beta vulgaris L. (Chenopodiaceae), assessed by RFLP and microsatellite markers. Theor Appl Genet 98:1194–1201
Feghhi SMA, Norouzi P, Saidi A, Zamani K, Amiri R (2012) Identification of SCAR and RAPD markers linked to Rz1 gene in Holly sugar beet using BSA and two genetic distance estimation methods. Electr J Plant Breed 3:598–605
Fénart S, Austerlitz F, Cuguen J, Arnaud J-F (2007) Long distance pollen-mediated gene flow at a landscape level: the weed beet as a case study. Mol Ecol 16:3801–3813
Fénart S, Arnaud JF, De Cauwer I, Cuguen J (2008) Nuclear and cytoplasmic genetic diversity in weed beet and sugar beet accessions compared to wild relatives: new insights into the genetic relationships within the Beta vulgaris complex species. Theor Appl Genet 116:1063–1077
Fievet V, Touzet P, Arnaud J-F, Cuguen J (2007) Spatial analysis of nuclear and cytoplasmic DNA diversity in wild sea beet (Beta vulgaris ssp. maritima) populations: do marine currents shape the genetic structure? Mol Ecol 16:1847–1864
Fischer HE (1989) Origin of the ‘Weisse Schlesische Rübe’ (white Silesian beet) and resynthesis of sugar beet. Euphytica 41:75–80
Frese L, Desprez B, Ziegler D (2001) Potential of genetic resources and breeding strategies for base-broadening in Beta. In: Cooper HD, Spillane C, Hodgkin T (eds) Broadening the genetic base of crop production. IPGRI/FAO, Rome, pp 295–309
Gidner S, Lennefors B-L, Nilsson N-O, Bensefelt J, Johansson E, Gyllenspetz U, Kraft T (2005) QTL mapping of BNYVV resistance from the WB41 source in sugar beet. Genome 48:279–285
Giorio G, Gallitelli M, Carriero F (1997) Molecular markers linked to rhizomania resistance in sugar beet, Beta vulgaris, from two different sources map to the same linkage group. Plant Breed 116:401–408
Grimmer MK, Trybush S, Hanley S, Francis SA, Karp A, Asher MJC (2007) An anchored linkage map for sugar beet based on AFLP, SNP and RAPD markers and QTL mapping of a new source of resistance to Beet necrotic yellow vein virus. Theor Appl Genet 114:1151–1160
Grimmer MK, Kraft T, Francis SA, Asher MJC (2008) QTL mapping of BNYVV resistance from the WB258 source in sugar beet. Plant Breed 127:650–652
Hjerdin A, Säll T, Nilsson NO, Bornman CH, Halldén C (1994) Genetic variation among wild and cultivated beets of the section Beta as revealed by RFLP analysis. J Sugar Beet Res 31:59–67
Jung C, Pillen K, Frese L, Fähr S, Melchinger AE (1993) Phylogenetic relationships between cultivated and wild species of the genus Beta revealed by DNA “fingerprinting”. Theor Appl Genet 86:449–457
Kadereit G, Hohmann S, Kadereit JW (2006) A synopsis of Chenopodiaceae subfam. Betoideae and notes on the taxonomy of Beta. Willdenowia 36:9–19
Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
Koo WW, Taylor RD (2012) 2012 outlook of the U.S. and world sugar markets, 2011–2021. Agribus Appl Econ Rep 692:1–29
Krichevsky A, Zaltsman A, King L, Citovsky V (2012) Expression of complete metabolic pathways in transgenic plants. Biotechnol Genet Eng Rev 28:1–13
Kubis S, Heslop-Harrison JS, Schmidt T (1997) A family of differentially amplified repetitive DNA sequences in the genus Beta reveals genetic variation in Beta vulgaris subspecies and cultivars. J Mol Evol 44:310–320
Lein JC, Asbach K, Tian Y, Schulte D, Li C, Koch G, Jung C, Cai D (2006) Resistance gene analogues are clustered on chromosome 3 of sugar beet and cosegregate with QTL for rhizomania resistance. Genome 50:61–71
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:381–398
Litwiniec A, Łukanowski A, Gośka M (2014) RNA silencing mechanisms are responsible for outstanding resistance of some wild beets against rhizomania. A preliminary evidence-based hypothesis. J Anim Plant Sci 21:3273–3292
McDermott JM, McDonald BA (1993) Gene flow in plant pathosystems. Annu Rev Phytopathol 31:353–373
McGrath JM, Derrico CA, Yu Y (1999) Genetic diversity in selected, historical US germplasm and Beta vulgaris ssp. maritima. Theor Appl Genet 98:968–976
Mita G, Dani M, Casciari P, Pasquali A, Selva E, Minganti C, Piccardi P (1991) Assessment of the degree of genetic variation in beet based on RFLP analysis and the taxonomy of Beta. Euphytica 55:1–6
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 176–192
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nishizawa S, Kubo T, Mikami T (2000) Variable number of tandem repeat loci in the mitochondrial genomes of beets. Curr Genet 37:34–38
Nouhi A, Amiri R, Haghnazari A, Saba J, Mesbah M (2008) Tagging of resistance gene(s) to rhizomania disease in sugar beet (Beta vulgaris L.). Afr J Biotechnol 7:430–433
Panella L, Lewellen RT (2007) Broadening the genetic base of sugar beet: introgression from wild relatives. Euphytica 154:382–400
Pelsy F, Merdinoglu D (1996) Identification and mapping of random amplified polymorphic DNA markers linked to a rhizomania resistance gene in sugar beet (Beta vulgaris L.) by bulked segregant analysis. Plant Breed 115:371–377
Reamon- Büttner SM, Wricke G, Frese L (1996) Interspecific relationship and genetic diversity in wild beets in section Corollinae genus Beta: isozyme and RAPD analyses. Genet Resour Crop Evol 43:261–274
Richards CM, Brownson M, Mitchell SE, Kresovich S, Panella L (2004) Polymorphic microsatellite markers for inferring diversity in wild and domesticated sugar beet (Beta vulgaris). Mol Ecol Notes 4:243–245
Saccomani M, Stevanato P, Trebbi D, McGrath JM, Biancardi E (2009) Molecular and morpho-physiological characterization of sea, ruderal and cultivated beets. Euphytica 169:19–29
Scholten OE, Klein-Lankhorst RM, Esselink DG, De Bock TSM, Lange W (1997) Identification and mapping of random amplified polymorphic DNA (RAPD) markers linked to resistance against beet necrotic yellow vein virus (BNYVV) in Beta accessions. Theor Appl Genet 94:123–130
Scholten OE, De Bock TSM, Klein-Lankhorst RM, Lange W (1999) Inheritance of resistance to beet necrotic yellow vein virus in Beta vulgaris conferred by a second gene for resistance. Theor Appl Genet 99:740–746
Scott HJ, Ford-Lloyd BV, Williams JT (1977) Patellifolia, nomen novum (Chenopodiaceae). Taxon 26:284
Smulders MJM, Esselink GD, Everaert I, De Riek J, Vosman B (2010) Characterisation of sugar beet (Beta vulgaris L. ssp. vulgaris) varieties using microsatellite markers. BMC Genet 11:41
Stevanato P, Trebbi D, Biancardi E, Cacco G, McGrath JM, Saccomani M (2013) Evaluation of genetic diversity and root traits of sea beet accessions of the Adriatic Sea coast. Euphytica 189:135–146
Thiel H, Hleibieh K, Gilmer D, Varrelmann M (2012) The P25 pathogenicity factor of Beet necrotic yellow vein virus targets the sugar beet 26S proteasome involved in the induction of a hypersensitive resistance response via interaction with an F-box protein. Mol Plant Microbe Interact 25:1058–1072
Van Geyt JPC, Lange W, Oleo M, De Bock ThSM (1990) Natural variation within the genus Beta and its possible use for breeding sugar beet: a review. Euphytica 49:57–76
Viard F, Bernard J, Desplanque B (2002) Crop-weed interactions in the Beta vulgaris complex at a local scale: allelic diversity and gene flow within sugar beet fields. Theor Appl Genet 104:688–697
Viard F, Arnaud J-F, Delescluse M, Cuguen J (2004) Tracing back seed and pollen flow within the crop-wild Beta vulgaris complex: genetic distinctiveness vs. hot spots of hybridization over a regional scale. Mol Ecol 13:1357–1364
Winter P, Kahl G (1995) Molecular marker technologies for plant improvement. World J Microbiol Biotechnol 11:438–448
Acknowledgments
The authors would like to express their gratitude to Agnieszka Żukowska and Dariusz Pianka, Bio-Rad Poland Ltd., for their support, to Agnieszka Chrustek and Żaneta Świtalska, graduate volounteers at the Laboratory of Biotechnology for their technical assistance, as well as to the representatives of the Polish breeding company of sugar beet Kutnowska Hodowla Buraka Cukrowego Ltd., in Straszków for cooperation. The authors appreciate kind suggestions of Lothar Frese, Crop Expert and Database Manager at International Data Base for Beet as to selection of some valuable accessions for rhizomania resistance studies. The study was supported by the funds of the State budget of Poland, the Ministry of Science and Higher Education for the Plant Breeding and Acclimatization Institute–NRI, Project No. 1-4-01-4-02.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Litwiniec, A., Gośka, M., Choińska, B. et al. Evaluation of rhizomania-resistance segregating sequences and overall genetic diversity pattern among selected accessions of Beta and Patellifolia. Potential implications of breeding for genetic bottlenecks in terms of rhizomania resistance. Euphytica 207, 685–706 (2016). https://doi.org/10.1007/s10681-015-1570-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1570-5