Abstract
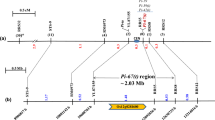
In the present study, we performed the resistance assessment by rice blast inoculation on IRBLta2-Re and IRBLta-CP1, the experimental lines supposed to carry rice blast resistance genes Pita2 and Pita, respectively. The analysis by using 196 rice blast isolates derived from China indicated that the resistance spectrum of IRBLta2-Re was broader than that of IRBLta-CP1. Both IRBLta2-Re and IRBLta-CP1 have the Pita gene by analyzing the functional single amino acid difference of Pita/pita locus. To identify the additional gene in IRBLta2-Re, 1250 F2 individuals from the cross between CO39 and IRBLta2-Re were used as the mapping population. The F2 population was inoculated with the blast isolate 08-T4 which was incompatible to IRBLta2-Re, but compatible to CO39 and IRBLta-CP1. In the phenotypic data analysis, the F2 population segregated in a 3:1 ratio for resistant and susceptible plants, respectively, suggesting that IRBLta2-Re has an additional resistance gene other than Pita, which was tentatively designated Pita3(t) (supposed to be Pita2). To identify the Pita3(t), a total of 50 microsatellite and 12 position specific microsatellite markers distributed by two sides of the Pita gene were selected in the parent polymorphism screening. The results showed that PT4 and PT5 were co-segregated with the target gene. A contig map corresponding to the resistance gene and Pita genes was constructed based on the fine mapping and bioinformatics assay. The resistance gene, Pita3(t), was, thus, assumed to be in an interval of approximately 178 kb which containing a total of 5 NBS–LRR genes, and was about 500 kb away from the Pita gene.



Similar content being viewed by others
References
Ashikawa IN, Hayashi H, Yamane H, Kanamori H, Wu J, Matsumoto T, Ono K, Yano M (2008) Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180:2267–2276
Bryan GT, Wu KS, Farrall L, Jia YL, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12:2033–2045
Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, Ma B, Wang Y, Zhao X, Li S, Zhu L (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804
Chen S, Huang ZH, Zeng LX, Yang JY, Liu QG, Zhu XY (2008) High resolution mapping and gene prediction of Xanthomonas oryzae pv. oryzae resistance gene Xa7. Mol Breed 22:433–441
Das A, Soubam D, Singh PK, Thakur S, Singh NK, Sharma TR (2012) A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct Integr Genomics 12:215–228
DeYoung BJ, Innes RW (2006) Plant NBS–LRR proteins in pathogen sensing and host defense. Nat Immunol 7:1243–1249
Fjellstrom R, Conaway-Bormans CA, McClung AM, Marchetti MA, Shank AR, Park WD (2004) Development of DNA markers suitable for marker assisted selection of three Pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci 44:1790–1798
Fukuoka S, Shimizu T, Saka N, Koga H, Ebana K, Hirochika H, Yano M, Okuno K (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 21(325):998–1001
Gupta SK, Rai AK, Kanwar SS, Chand D, Singh NK, Sharma TR (2012) The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J Exp Bot 63:757–772
Hayashi K, Yoshida H (2009) Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J 57:413–425
Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, Yano M, Takatsuji H (2010) Durable panicle blast-resistance gene Pb1 encodes an atypical CC–NBS–LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J 64:498–510
Hua L, Wu J, Chen C, Wu W, He X, Lin F, Wang L, Ashikawa I, Matsumoto T, Wang L, Pan Q (2012) The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet 125:1047–1055
Huang D, Qiu Y, Zhang Y, Huang F, Meng J, Wei S, Li R, Chen B (2012) Fine mapping and characterization of BPH27, a brown planthopper resistance gene from wild rice (Oryza rufipogon Griff.). Theor Appl Genet 126:219–229
Jia YL, Jia MH, Wang XY, Liu GJ (2012) Indica and japonica crosses resulting in linkage block and recombination suppression on rice chromosome 12. PLoS One 7:1–7
Kiyosawa S (1971) Genetical approach to the biochemical nature of plant disease resistance. Japan Agric Res Quart 6:72–80
Kiyosawa S, Mackill DJ, Bonman JM, Tanaka Y, Ling ZZ (1986) An attempt of classification of world’s rice varieties based on reaction pattern to blast fungus strains. Bull Natl Inst Agrobiol Resour 2:13–39
Laitinen RA, Schneeberger K, Jelly NS, Ossowski S, Weigel D (2010) Identification of a spontaneous frame shift mutation in a nonreference Arabidopsis accession using whole genome sequencing. Plant Physiol 153:652–654
Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP, Yi G, Roh JH, Lee S, An G, Hahn TR, Wang GL, Ronald P, Jeon JS (2009) Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two CC-NB-LRR genes. Genetics 181:1627–1638
Lin F, Chen S, Que Z, Wang L, Liu X, Pan Q (2007) The blast resistance gene Pi37 encodes a nucleotide binding site leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 177:1871–1880
Liu XQ, Lin F, Wang L, Pan QH (2007) The in silico map-based cloning of Pi36, a rice CC–NBS–LRR gene which confers race specific resistance to the blast fungus. Genetics 176:2541–2549
Liu J, Wang X, Mitchell T, Hu Y, Liu X, Dai L, Wang GL (2010) Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction. Mol Plant Pathol 11:419–427
McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
Monosi B, Wisser RJ, Pennill L, Hulbert SH (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109:1434–1447
Murray MG, Thompson WK (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res 8:4321–4325
Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Takahiro Fujibe T, Matsumura H, Shenton M, Galam D, Undan J, Ito A, Sone T, Terauchi R (2011) A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS–LRR protein genes. Plant J 66:467–479
Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Dai L, Han B, Wang GL (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172:1901–1914
Rybka K, Miyamoto M, Ando I, Saito A, Kawasaki S (1997) High resolution mapping of the indica-derived rice blast resistance genes II. Pi-ta 2 and Pi-ta and a consideration of their origin. Mol Plant Microbe Interact 10:517–524
Shang JJ, Tao Y, Chen XW, Zou Y, Lei CL, Wang J, Li XB, Zhang MJ, Zhao XF, Lu ZK, Xu JC, Cheng ZK, Wan JM, Zhu LH (2009) Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site-leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182:1303–1311
Sharma TR, Madhav MS, Singh BK, Shanker P, Jana TK, Dalal V, Pandit A, Singh A, Gaikwad K, Upreti HC, Singh NK (2005) High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to Magnaporthe grisea. Mol Genet Genomics 274:569–578
Takahashi A, Hayashi N, Miyao A, Hirochika H (2010) Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC Plant Biol 10:175
Tsunematsu H, Yanoria MJT, Ebron LA, Hayashi N, Ando I, Kato H, Imbe T, Khush GS (2000) Development of monogenic lines for rice blast resistance. Breed Sci 50:229–234
Uchida N, Sakamoto T, Kurata T, Tasaka M (2011) Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol 52:716–722
Wang GL, Mackill DJ, Michael Bonman JM, McCouch SR, Champoux MC, Nelson RJ (1994) RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 136:1421–1434
Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T (1999) The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J 19:55–64
Wang Y, Wang D, Deng X, Liu J, Sun P, Liu Y, Huang H, Jiang N, Kang H, Ning Y, Wang Z, Xiao Y, Liu X, Liu E, Dai L, Wang GL (2012) Molecular mapping of the blast resistance genes Pi2-1 and Pi51(t) in the durably resistant rice ‘Tianjingyeshengdao’. Phytopathology 102:779–786
Yang JY, Chen S, Zeng LX, Li YL, Chen Z, Li CY, Zhu XY (2008) Race specificity of major rice blast resistance genes to Magnaporthe grisea isolates collected from indica rice in Guangdong, China. Rice Sci 15:311–318
Yuan B, Zhai C, Wang W, Zeng X, Xu X, Hu H, Lin F, Wang L, Pan Q (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC–NBS–LRR genes. Theor Appl Genet 122:1017–2108
Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, Wang L, Pan Q (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189:321–334
Zhang QF, Shen BZ, Dai XK, Mei MH, Saghai-Maroof MA, Li ZB (1994) Using bulked extremes and recessive class to map genes for photoperiod-sensitive genetic male sterility in rice. Proc Natl Acad Sci USA 91:8675–8679
Zhou B, Qu S, Liu G, Dolan M, Sakai H, Lu G, Bellizzi M, Wang GL (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe Interact 19:1216–1228
Zhu X, Chen S, Yang J, Zhou S, Zeng L, Han J, Su J, Wang L, Pan Q (2012) The identification of Pi50(t), a new member of the rice blast resistance Pi2/Pi9 multigene family. Theor Appl Genet 124:1295–1304
Acknowledgments
This research was supported by the National Natural Science Foundation (31101403, U1031002), the National Transgenic Research Projects (2009ZX08009-006B),the Earmarked Fund for Modern Agro-Industry Technology Research System (CARS-01-24, Yuecaijiao [2009] No. 356), Commonweal Specialized Research Fund of China Agriculture (201203014), the Science and Technology Projects of Guangzhou (2012J4300059, 2012J2200066), and the President Science Foundation of Guangdong Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shen Chen and Jing Su are contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, S., Su, J., Han, J. et al. Resistance spectrum assay and fine mapping of the blast resistance gene from a rice experimental line, IRBLta2-Re. Euphytica 195, 209–216 (2014). https://doi.org/10.1007/s10681-013-0992-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-013-0992-1