Abstract
Biosecurity strategies that aim to restrict the spread of invasive pests can benefit from knowing where incursions have come from and whether cryptic establishment has taken place. This knowledge can be acquired with genomic databanks, by comparing genetic variation in incursion samples against reference samples. Here we use genomic databanks to characterise incursions of two mosquito species within Australia, and to observe how genomic tracing methods perform when databank samples have limited genetic differentiation and were collected tens of generations ago. We used a deep learning method to trace a 2021 invasion of Aedes aegypti in Tennant Creek, Northern Territory, to Townsville, Queensland, and to trace two years of Ae. albopictus incursions to two specific islands in the Torres Strait. Tracing had high precision despite 30–70 generations separating incursion and reference samples, and cross-validation of reference samples assigned them to the correct origin in 87% of cases. Similar precision was not achieved with PCAs, which performed particularly poorly for tracing when the invasion had been subject to strong drift effects. Targeted assays also provided additional information on the origin of the Tennant Creek Ae. aegypti, in this case by comparing Wolbachia infection data and mitochondrial DNA variation. Patterns of relatedness and inbreeding indicated that Tennant Creek was likely invaded by one family of Ae. aegypti, while Torres Strait incursions were independent and indicated no cryptic establishment. Our results highlight the value of genomic databanks that remain informative over years and for a range of biological conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Invasions of exotic pests confer a global economic burden of over $200B annually (Diagne et al. 2021). These costs have risen in recent decades and are expected to rise further as invasions continue to increase in frequency worldwide (Sardain et al. 2019). Invasive insect pests mostly establish via passive movement on ships, planes, or land vehicles, and are typically difficult to eradicate with insecticides due to resistance that is either present on arrival or evolves shortly afterwards (Gao et al. 2021). Border biosecurity operations have demonstrated success at restricting the spread of pests (Caley et al. 2015; Muzari et al. 2017), however, resource limitations constrain the number of inspections on arriving vessels and the number of border sections that can be effectively monitored.
Biosecurity operations can be greatly assisted by knowledge of pest incursion pathways, as this allows for more frequent inspection of goods and conveyances from likely source locations (Robinson et al. 2011) and regular inspection of known entry points (Myers et al. 2000; Mehta et al. 2007). While pathways can be easily inferred if incursions are detected on their vessel of entry, incursions that are detected away from vessels pose a greater challenge. Moreover, it is not always clear if these individuals are from the original incursions or from cryptically established invasive populations, the latter of which presents a new problem requiring a shift in management strategy. These biosecurity issues link to current areas of research in pest ecology, such as inferring the direction of passive movement (Hulme et al. 2008), changes in movement vectors over time (Zeigler and Fagan 2014), establishment likelihood as a function of the size and frequency of introductions (i.e. propagule pressure) (Stringham and Lockwood 2021), and population growth rates following establishment (Tobin et al. 2011). Knowledge of passive movement pathways, propagule pressure, and cryptic establishment can help inform border biosecurity programs such as the program that has stopped the Asian tiger mosquito Aedes albopictus from invading the Australian mainland for 18 years so far (Muzari et al. 2017).
Genomic databanks are an emerging technology that show promise for investigating these processes. When databanks contain samples covering a sufficient geographical range, they can indicate specific movement of incursive pests across international borders (Schmidt et al. 2019, 2020b; Popa-Báez et al. 2021; Parvizi et al. 2023) and among geographical regions (Chen et al. 2021; Schmidt et al. 2021c). Rather than inferring the origin of established invasive populations, which can certainly be achieved with such databanks (Gloria-Soria et al. 2018; Sherpa et al. 2019; Schmidt et al. 2020a; Kelly et al. 2021; Yan et al. 2021), these emerging methods apply pairwise kinship tests or assignment tests to incursive individuals directly. Genomic signals of inbreeding and kinship patterns within and among the incursive individuals may also provide further insight into incursion size and cryptic establishment.
While pest management is beginning to employ genomic databanks, studies have yet to evaluate their power to infer specific movement when reference samples are genetically similar across space, as is common in pest populations (Fitzpatrick et al. 2012), or when tens of generations of drift and migration have occurred since sampling. These questions are vital for the wider uptake of these methods as they determine the size of the initial sequencing investment and the length of time a databank will remain representative of extant populations. This study considers these questions directly in an analysis of two globally invasive mosquito species, the dengue mosquito, Aedes aegypti and the tiger mosquito, Ae. albopictus. Both species are readily transported by human conveyances including boats and planes, and genomic studies of recent invasions have found these are often linked to multiple geographically distinct source locations (Schmidt et al. 2021a). Here we focus on recent detections of these species within Australia, away from entry ports and of unknown provenance.
For Ae. aegypti, we trace an invasion of Tennant Creek, Northern Territory, that was detected in early 2021 and established rapidly across the town. This species had previously been eliminated from Tennant Creek in 2006, following its establishment in 2004, and again in 2014 following its establishment in 2011 (Pettit and Kurucz 2014; Whelan et al. 2020). An analysis of variation in the mtDNA COI gene concluded that the 2004 incursion had most likely originated from Cairns or somewhere nearby, and had probably been transported into Tennant Creek as eggs in dry receptacles (Beebe et al. 2005; Whelan et al. 2012). Both elimination programs were funded in collaboration between the Northern Territory Government and the Commonwealth Department of Health to prevent the establishment of Ae. aegypti in the Northern Territory and subsequently in Western Australia.
For Ae. albopictus, we trace a set of recent incursions detected in the Torres Strait, Queensland, on islands where this species had been successfully eliminated (Muzari et al. 2017). The Torres Strait invasion of this species began in 2004 (Ritchie et al. 2006) and has since been shown to have a predominantly Indonesian genetic background (Beebe et al. 2013; Schmidt et al. 2021c). This background is distinct from other Ae. albopictus including those from nearby Papua New Guinea, except for the South Fly District, which may have been invaded from the same source as the Torres Strait (Maynard et al. 2017). Since 2008, a cordon sanitaire biosecurity strategy was imposed to regulate the movement of people and goods between the invaded islands of the Torres Strait and the uninvaded southern islands, which has so far successfully stopped the spread of Ae. albopictus to the Australian mainland (Muzari et al. 2017). Islands south of the cordon-sanitaire undergo periodical surveillance for Ae. albopictus incursions, and a previous study of this region linked three 2019 incursions to two islands, Keriri and Moa (Schmidt et al. 2021c).
In testing these ideas, we show that the deep learning method Locator (Battey et al. 2020) provides remarkably high levels of unambiguous and unbiased assignment during cross-validation of reference samples. This method also provides inferences of passive movement through precise tracing of incursion samples to their geographical origins, despite high genetic similarity, strong effects of drift, and tens of generations of evolution separating incursions and reference samples. In the case of Ae. aegypti, precision in tracing was further improved by targeted assays for insecticide resistance and a Wolbachia bacterial infection, which follow releases to establish Wolbachia in north-eastern Australia over the last decade (Schmidt et al. 2017; O’Neill et al. 2019; Ryan et al. 2020). Finally, by investigating genomic patterns within and between incursive individuals, we show that the Ae. aegypti invasion was likely sourced from a single geographical location and potentially from a single family, while the Ae. albopictus incursions were independent and showed no signs of cryptic establishment.
Materials and methods
Mosquito sample collection
Aedes aegypti invasion of Tennant Creek, NT, Australia
Aedes aegypti was detected in Tennant Creek during routine surveillance between 22nd and 26th February 2021. Four adult mosquitoes from this invasion (three females, one male) were collected from a single property on 26th February. Follow-up sampling between 9th and 11th March located an additional five adults (three females, two males) and nine immatures at four properties within ~ 80 m of the original detection. After sampling, the invasion was found to have established across Tennant Creek.
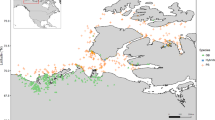
To trace the source of this invasion, we built a genomic databank from 80 Ae. aegypti sampled from 11 locations across its Australian distribution in Queensland, from the northern islands of the Torres Strait (9.3° S) to Goomeri in southeast Queensland (26.2° S) (Fig. 1a, b). These were sampled between 2009 and 2017. We also included previously-generated sequence data from Cairns and Townsville. Both of these cities have been subject to Wolbachia mosquito releases for dengue transmission control, with Cairns releases beginning in 2013 (Schmidt et al. 2017) and Townsville releases in 2014 (O’Neill et al. 2019). The Cairns sample was taken in 2015 after releases (Schmidt et al. 2018), and included some individuals infected with Wolbachia, whereas the Townsville sample was taken in 2013 before releases (Rašić et al. 2016). The Townsville sample thus represents a pre-Wolbachia snapshot of this population.
Distributions (a, c) and sample locations (b, d) of Ae. aegypti (a, b) and Ae. albopictus (c, d). Black squares indicate current Australian distributions based on Atlas of Living Australia records. Blue circles show locations of reference samples. In (b), the orange square indicates Tennant Creek. In (d), the orange square indicates Thursday Island and the red square indicates Horn Island. Maps use shapefiles made available by www.naturalearthdata.com (a–c) and the Torres Strait Clear sky Landsat (d; https://eatlas.org.au/data/uuid/71c8380e-4cdc-4544-98b6-8a5c328930ad)
Aedes albopictus incursions in the Torres Strait Islands, QLD, Australia
Aedes albopictus incursions were detected during routine surveillance on two islands in the Torres Strait where the species had been successfully eliminated (Muzari et al. 2017). Surveillance conducted in January and February 2021 detected four adult individuals on Thursday Island and three on Horn Island (Fig. 1c, d). Surveillance in January 2022 detected a further four adults on Thursday Island and five on Horn Island. In both years and on both islands, incursions were detected across a wide geographical range (Figures S1–4). There were no further detections of Ae. albopictus on either island over the periods March–December 2021 and February–June 2022. This may reflect a failure of Ae. albopictus to establish on these islands, but these observations also fit with cryptic establishment at low frequencies.
To trace the source of these incursions, we used a genomic databank containing previously generated sequence data of 240 Ae. albopictus from 12 Torres Strait Island communities across 11 islands, collected in 2018 (Schmidt et al. 2021c) (Fig. 1c, d). These samples were individually georeferenced and were collected from across each island.
DNA sequencing and bioinformatic processing
Genomic DNA was extracted from the 98 Ae. aegypti and 16 Ae. albopictus using Qiagen DNeasy® Blood & Tissue Kits (Qiagen, Hilden, Germany). Extracted DNA was used to build double digest restriction-site associated DNA (ddRAD; Peterson et al. (2012)) sequencing libraries following the protocol of Rašić et al. (2014). A technical replicate of one of the Torres Strait Island incursion samples from Thursday Island, TI21-1, was taken through the entire protocol after DNA extraction. Libraries were individually barcoded, then sequenced on a HiSeq 4000 using 150 bp chemistry.
New and old sequence data were combined for each species and run through the same bioinformatic pipeline. We used the Stacks v2.54 (Rochette et al. 2019) program process_radtags to demultiplex sequences, trim them to 140 bp and remove sequences with Phred scores below 20. As the Cairns and Townsville libraries were produced using 100 bp chemistry, we trimmed these to 80 bp. Sequences were aligned to the nuclear genome assembly for Ae. aegypti, AaegL5 (Matthews et al. 2018), and the linkage-based assembly for Ae. albopictus, AalbF3 (Boyle et al. 2021). Alignment was in Bowtie2 v2.3.4.3 (Langmead and Salzberg 2012) using “–very-sensitive” settings. Sequences were built into catalogs for each species using the Stacks program ref_map. The Stacks program “populations” was used to export VCF files containing SNP genotypes for all individuals in each catalog, filtering to retain SNPs called in at least 50% of individuals from each population and 90% of individuals total, and with a minor allele count ≥ 3 (Linck and Battey 2019). Two of the 18 Tennant Creek Ae. aegypti and 1 incursive Ae. albopictus from Thursday Island in 2022 had too much missing data for tracing (> 60%) and were omitted. We also omitted ten Ae. aegypti and three Ae. albopictus reference samples with > 30% missing data. All other samples had < 30% missing data.
Cross-validation and filtering of reference datasets
To assess the robustness of the reference datasets we performed cross-validation. Each individual in turn was omitted from the training set and treated as an individual of unknown location, and then Locator (Battey et al. 2020; see below) was used to infer the geographical origin of the individual. Assignments were assessed qualitatively for accuracy (i. e. is its origin unambiguous?) as well as quantitatively for precision (i. e. how far from its true origin?). For the Ae. aegypti dataset, we treated nearby samples from Bluff, Capella, and Emerald (QLD, max separation 120 km) as one location, as well as combining those from Duaringa, Mt Morgan and Rockhampton (QLD, max separation 95 km) and those from Gin Gin and Goomeri (QLD, separation 133 km). This was because sample sizes were small (5) from each specific location. Cross-validation was also used to test two site filtering protocols: where SNPs were not filtered for linkage; and where SNPs within 25 kbp of other SNPs were filtered.
Incursion tracing
Genome-wide genetic variation
We used Locator (Battey et al. 2020) to infer the origin of the 16 invasive Ae. aegypti and the 16 incursive Ae. albopictus (including the technical replicate). Locator is a deep learning method that uses individual genotypes of known geographical location (i. e. the reference individuals) to predict the locations of a set of individuals of unknown location (i. e. the incursive mosquitoes). For each incursive mosquito, we set Locator to provide a point estimate of its origin and 1,000 bootstrap subsamples to indicate confidence around these estimates.
We benchmarked Locator’s performance against inferences derived from principal components analysis (PCA), a common tool for investigating genomic similarity among populations. PCAs were run in the R package LEA (Frichot and François 2015), using samples in this study and others from the Indo-Pacific region previously published in Schmidt et al. (2020a, 2021c).
Wolbachia infection
The Tennant Creek Ae. aegypti were assessed for infection of wMel Wolbachia, which are present at high frequency in Ae. aegypti populations in Cairns, Townsville, and several other locations in northern Queensland, but not in central Queensland populations. Wolbachia infection was assessed using a high-throughput PCR assay (Lee et al. 2012) which was run twice on all 18 samples.
In the case of uninfected samples, we also considered the possibility that the wMel infection was initially present but then lost from the invasive population due to the vulnerability of this Wolbachia strain to high temperatures (Ross et al. 2017) that are common in Tennant Creek. As Wolbachia is coinherited with mtDNA through the maternal line (Hoffmann and Turelli 1997), wMel-infected individuals will all have inherited the same mtDNA haplotype from the same common ancestor within the past ~ 10 years. This mtDNA haplotype will be retained even if Wolbachia has been lost (Yeap et al. 2016), and its presence would be revealed by high genetic similarity in mtDNA between the Tennant Creek Ae. aegypti and samples infected with wMel.
To test this hypothesis, we looked at mtDNA variation among our Ae. aegypti samples alongside variation from 38 wMel-infected (Wol+) and 87 uninfected wildtype (Wol−) individuals from Cairns, sampled in 2015 (Schmidt et al. 2018). We aligned demultiplexed sequences to the Ae. aegypti mtDNA genome assembly (Behura et al. 2011) and called mtDNA SNPs using Stacks. We used VCFtools to output mtDNA genotypes for all individuals with minimum depth of 5, omitting SNPs with any heterozygous genotypes which could represent NUMTs (Hlaing et al. 2009).
Pyrethroid resistance mutations
The Tennant Creek Ae. aegypti were assessed for three pyrethroid resistance mutations, V1016G, F1534C, and S989P, which are common to Ae. aegypti across the Indo-Pacific (Endersby-Harshman et al. 2020), but not within Australia (Endersby-Harshman et al. 2017). Monitoring mosquito incursions for these mutations is standard practise to confirm susceptibility to insecticide-based control and to ensure these mutations are not introgressing through Australian Ae. aegypti as they have through other Indo-Pacific populations (Endersby-Harshman et al. 2020). For each individual, we ran three replicates of the Custom TaqMan® SNP Genotyping Assays (Life Technologies Corporation, Carlsbad, CA, USA) described in Endersby-Harshman et al. (2020), using a LightCycler II 480 (Roche, Basel, Switzerland) real time PCR machine in a 384-well format.
Genetic relatedness and inbreeding in the incursions
We investigated patterns of genetic relatedness (k) among pairs of individuals (dyads) in the Tennant Creek and Torres Strait incursions, and calculated Wright’s inbreeding coefficient (F) for every individual. The patterns can help indicate whether the samples at either location had arrived as part of a single incursion (i.e. on a single vessel or set of vessels at a single time point) or as several distinct incursions, and whether cryptic establishment has taken place.
For instance, if specific incursive individuals have higher k scores than other dyads in the sample, these are likely to be close kin, and as mosquitoes have very large population sizes it is most likely that they are related offspring of a single female that has laid eggs after the incursion or on a vessel that transports the incursion. If incursive individuals have higher F scores than other individuals, they are likely to be the product of inbreeding following a single incursion of one or a few individuals. High F scores can thus also indicate cryptic establishment if incursion samples are inbred but no established invasion is apparent.
We used Loiselle’s k (Loiselle et al. 1995) to estimate genetic relatedness, calculated in SPAGeDi (Hardy and Vekemans 2002) using the linkage-thinned datasets for each species. Loiselle’s k describes correlations in the frequencies of homologous alleles, summarised across the genome (Loiselle et al. 1995). We used this estimator as it makes no assumptions regarding F and is suitable for markers with low polymorphism (Vekemans and Hardy 2004). We calculated F in VCFtools (Danecek et al. 2011). As estimators of individual-level variation can be biased by uneven sample size and missing data (Schmidt et al. 2021b), we subsampled and refiltered each databank before calculating F. For Ae. aegypti, we used the Tennant Creek (n = 16), Townsville (n = 16) and Cairns (n = 15) samples. For Ae. albopictus, we used n = 15 samples from each reference population and the n = 15 incursion samples. We refiltered to omit all sites with any missing genotypes (–max-missing 1) and to keep only sites with mean depth ≥ 20 (–min-meanDP 20), and did not filter singletons and doubletons.
Results
Cross-validation and filtering of reference datasets
Cross-validation indicated that different filtering settings were optimal for each species (Fig. 2). In Ae. aegypti, 4.9% (5 out of 102) of reference samples were assigned to the incorrect origin when SNPs were thinned for linkage, and 6.9% (7 out of 102) without thinning. Unambiguous assignment of a sample to its true origin occurred in 88% of samples (thinned) or 76% of samples (no thinning). The average error was 47.1 km (thinned) or 60.7 km (no thinning), out of a maximum distance between samples of 1881.9 km.
In Ae. albopictus, 10.5% (25 out of 237) of samples were assigned to the incorrect origin for both thinning treatments. Unambiguous assignment occurred in 84% of samples (thinned) or 86% of samples (no thinning). One location, Badu Island, performed particularly poorly in the thinning treatment, with only 30% of individuals unambiguously assigned compared to 60% without thinning. Average error was 11.6 km (thinned) or 11.2 km (no thinning), out of a maximum distance of 213.4 km. There was no evidence in either species that incorrect assignments were biased to specific locations. We used the thinned dataset for Ae. aegypti tracing and the unthinned dataset for Ae. albopictus tracing, the latter of which was chosen due to the poor assignment to Badu Island with the thinned dataset.
Incursion tracing
Aedes aegypti invasion of Tennant Creek, NT, Australia
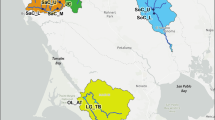
Genomic tracing with Locator was run sequentially on different subsets of samples. An initial run with all 102 reference samples and 11,866 thinned SNPs indicated all 16 samples had a likely origin between Cairns (16.9° S) and Rockhampton (23.4° S) (Figure S5). Rerunning Locator without reference samples outside this range and 11,495 SNPs indicated a likely origin of Townsville (Fig. 3). We checked whether this assignment to origin was biased by Townsville’s geographically central location by rerunning Locator using either Townsville and Cairns only (Figure S6) or Townsville and the southern samples only (Figure S7). In each case, incursives were still placed around Townsville, but Townsville and Cairns were slightly harder to differentiate.
Origins of the 16 Tennant Creek individuals inferred from genome-wide genetic variation. Blue circles indicate reference sample locations, red circles show inferred origins from 1000 bootstrap replicate runs of Locator. Figures S5–7 show results of similar analyses using the whole dataset (S5) or different subsets of the dataset (S6–7)
Assays for wMel Wolbachia indicated none of the Tennant Creek samples were infected with wMel, in contrast to positive controls from an infected line. Analysis of 116 mitochondrial SNPs showed that pairwise FST between Tennant Creek and Townsville was lower (FST = 0.24) than between Tennant Creek and either the wMel infected or uninfected Cairns samples (both FST = 0.85) (Table 1). This indicates that no wMel infection was ever present in the Tennant Creek samples or their maternal ancestors, otherwise the Tennant Creek sample should be more similar to the Cairns samples given that Cairns provided the genetic background for the original wMel infected strain (Walker et al. 2011). In the pyrethroid resistance assays, none of the three resistance alleles was present in the Tennant Creek samples, consistent with the absence of resistance alleles in North Queensland populations (Endersby-Harshman et al. 2017).
Aedes albopictus incursions in the Torres Strait Islands, QLD, Australia
Locator was run on Ae. albopictus using 73,685 SNPs. Differences in tracing outcomes were evident between the 2021 and 2022 incursions (Fig. 4), and these were not due to missing data which was similarly low in both years. For the 2021 samples, the only incursive individual that could be assigned to a specific location at high confidence was the Thursday Island sample TI21-1 and its technical replicate, assigned to the St Pauls community on Moa Island. Several of the other 2021 incursives had a likely origin in the Maluilgal (Western) island group, though there was considerable variation among bootstrap replicates. The 2022 samples showed much better assignment, with seven of the nine clearly linked to Keriri Island and six of these showing very low variation among bootstrap replicates. This included one incursive collected next to the airport (HI22-4). Keriri and Moa Island are the two islands to which incursions have previously been linked in this system (Schmidt et al. 2021c).
Origins of the 15 Torres Strait Islands individuals inferred from genome-wide genetic variation. Samples were detected on Horn Island (HI) or Thursday Island (TI) in either 2021 or 2022. A technical replicate (R) was made of the TI21-1 sample. Blue circles indicate reference sample locations, red circles show inferred origins from 1000 bootstrap replicate runs of Locator
Comparisons with PCA
PCA-based assessment of genetic clustering provided limited insight into the origin of the Ae. aegypti invasion (Fig. 5a–c). In each PCA, Tennant Creek Ae. aegypti were strongly differentiated from all other samples by variation on the PC1 axis. Tennant Creek did not cluster with any other population even at very broad scales, though were more closely linked with Australian samples than others from the Indo-Pacific (Fig. 5a).
PCAs of Ae. aegypti (a–c) and Ae. albopictus (d–f). PCAs show genetic relationships between incursions and global populations (a, d), Australian populations (b, c), Torres Strait Island clusters (e), and Torres Strait Islands (f). Islands that clustered apart from incursion samples in e were omitted for f. Island clusters in f are indicated in parentheses
Within Australia, Tennant Creek mosquitoes were closest to Cairns and Townsville (Fig. 5b), though when analysed against only these two populations they could not be linked to either (Fig. 5c). The possibility of the Tennant Creek invasion coming from an unsampled, highly differentiated population was very low, however; when analysed alongside Cairns and Townsville with the program Populations, Tennant Creek had far fewer private alleles than either Cairns or Townsville (Table 2). The proportion of polymorphic loci was likewise much lower in Tennant Creek. These findings strongly suggest that the differentiation of Tennant Creek on PC1 was not due to its genetic uniqueness but rather due to drift.
PCAs were able to link the Torres Strait incursions to the Torres Strait reference samples (Fig. 5d), though not at the specificity seen in the Locator analysis. PCA of the Torres Strait locations (Fig. 5e) and a subset of Torres Strait Islands that genetically overlapped the incursions in PC space (Fig. 5f) could not indicate specific islands of origin. Most of the incursion samples were placed near the origin, next to Keriri of the Inner Island cluster, but also overlapped with samples from other island clusters (Fig. 5e). The other two incursion samples (TI21-1 and its replicate, TI21-1r) were placed with the Moa Island populations of St Pauls and Kubin, which accords with Locator placing these samples in St Pauls (Fig. 5f, c.f. Fig. 4).
Genetic relatedness and inbreeding in the incursions
Figure 6 displays pairwise Loiselle’s k scores for the Tennant Creek (6a) and Torres Strait Island (6b) dyads in rank order, and Wright’s inbreeding coefficient (F) scores in the subsampled and refiltered datasets (5073 SNPs for Ae. aegypti (6c); 3223 SNPs for Ae. albopictus (6d)).
Genetic relatedness (k) and Wright’s inbreeding coefficients (F) for Ae. aegypti (a, c) and Ae. albopictus (b, d). Loiselle’s k scores are of dyads in Tennant Creek (a) and the Torres Strait Islands (b), where dark squares show correlations between homologous alleles in two different individuals and light squares show correlations within the same individual. F scores are of Ae. aegypti (c) and Ae. albopictus (d), where red squares indicate invasive individuals and grey circles indicate other individuals
Loiselle’s k scores represent either correlations between homologous alleles in two different individuals (dark squares) or the same individual (light squares). The relative magnitudes of k scores within and between individuals were different in the two datasets. In the Torres Strait Islands, k scores within individuals were all higher than the k scores between individuals (Fig. 6b). This pattern means that the two chromosomes within each individual are more genetically similar to each other than to chromosomes found in other individuals. The higher k within individuals suggests that none of the Torres Strait Island incursives are likely to be from the same family, as dyads from the same family would have chromosomes with high genetic similarity via identity by descent. Instead, these results are evidence that the 15 Torres Strait Island incursives each represent distinct incursions. This pattern was not observed at Tennant Creek, where k scores within individuals were mostly undifferentiated from and in many cases lower than k scores between individuals (Fig. 6a). Chromosomes within individuals are thus no more genetically similar than they are to chromosomes in other individuals. This suggests that the 16 Tennant Creek individuals are all likely to be from the same exact geographical location and are potentially all from the same family.
The above inferences were further supported by variation in F scores. In Tennant Creek, F scores indicated much higher inbreeding relative to the Cairns and Townsville reference individuals, suggesting that the Tennant Creek invasion was sourced from a single family that had been inbreeding following establishment (Fig. 6c, red squares). In the Torres Strait Islands, F scores in incursive individuals (Fig. 6d) were of similar magnitude to reference individuals, suggesting that the incursive mosquitoes were not from lineages that had been recently inbreeding.
Discussion
Genomic data allow for powerful analyses of pests, including detecting specific movement of individuals between invaded regions and into new regions. These movement inferences relate directly to organismal life histories and thus provide a means of using genetic data in ecological contexts, such as understanding passive movement (Hulme et al. 2008), how movement vectors change over time (Zeigler and Fagan 2014), and the extent of passive movement required for establishment (Stringham and Lockwood 2021). This study has demonstrated the use of a deep learning program, Locator (Battey et al. 2020), to infer geographical origins of invasive Aedes mosquitoes and, in doing so, infer the recent movement of specific individuals. Also, by comparing genetic structure within individuals, we found high levels of inbreeding in a recently-established Ae. aegypti invasion, which suggests that the Ae. aegypti invasion was sourced from one location and from a small genetic pool (e. g. a few individuals or a larger group of related individuals). There were no elevated levels of inbreeding in the Ae. albopictus incursions, suggesting that no cryptic establishment of Ae. albopictus has taken place in the Torres Strait.
As well as answering these specific biosecurity questions, this study helps clarify some broader ideas around the use of genomic databanks in pest management. Most importantly, we have shown via cross-validation that the databanks used in this study are sufficiently powered for precise and unbiased tracing, and our incursion tracing results confirm that this power remains high despite tens of generations of evolution and low geographical genetic structure. If sequences that adequately represent an individual’s true population are not included in the reference dataset, Locator tends to place the individual around the centre of the sampling range (Battey et al. 2020). This was not observed here, as the Tennant Creek invasion was traced to Townsville irrespective of its relative position in the sampling range (Figures S6–7) and the Torres Strait incursives were traced to specific islands in the Western edge of the range (Fig. 4). As several Torres Strait incursives were not traceable to any specific island, it is possible that these are from an unsampled location with a similar genetic background such as South Fly District in Papua New Guinea or an unsampled location in the Torres Strait.
Cross-validation also indicated different optimal filtering settings for the two databanks. While Locator’s developers have suggested to subsample bootstrap replicates from a set of unlinked SNPs (Battey et al. 2020), we found that thinning led to slightly worse cross-validation in Ae. albopictus, though in Ae. aegypti the unthinned dataset performed much worse (Fig. 2). This result may be due to the smaller amount of genetic variation in the Ae. albopictus databank, which when thinned was reduced to only 4,147 SNPs, as opposed to 14,648 for Ae. aegypti. Whole genome databanks would presumably provide much more genetic variation for tracing, but for species with large genomes the larger sequencing effort required for these may have to be offset by smaller sample sizes. Thankfully, whole genome data can be downsampled for compatibility with reduced-representation databanks, which is unfortunately not the case for reduced-representation data produced with different restriction enzymes which currently prohibits analysis of different global datasets (Schmidt et al. 2021a).
For databanks to provide value for money, they should be reusable for tracing over years or decades. However, genetic drift and dispersal will ensure that reference samples in the databank are increasingly less representative of extant populations with each passing generation. Our analysis of Ae. aegypti showed that the 2021 Tennant Creek invasion samples could be traced to the Townsville genetic background represented by samples collected in January 2014. Assuming 10 generations/year for Aedes, the Tennant Creek and Townsville samples should be separated by ~ 70 generations of evolution, plus several generations of strong genetic drift between invasion and detection. The databank of Torres Strait Ae. albopictus showed similar robustness in tracing, wherein a majority of the incursives detected 3–4 years (~ 30–40 generations) after the reference samples could be traced to two specific islands with very high confidence (Fig. 4). This is particularly noteworthy given the much finer spatial scale of the Torres Strait invasion and the low genetic differentiation among islands (FST ≈ 0.03, Schmidt et al. (2021c)). Differentiation between Cairns and Townsville Ae. aegypti is similarly very low (FST′ = 0.018, Schmidt et al. (2020a)) but assignments were unambiguous (Figs. 3, S6).
Using targeted assays for Wolbachia infection and pyrethroid resistance, combined with mtDNA data from the databank, we were able to increase the confidence of the origin of Tennant Creek Ae. aegypti over using nuclear genomic data alone. These findings indicate that the invasion may have been sourced from an unsampled location near to Townsville or from a region of outer Townsville where the wMel infection is uncommon, as wMel has remained close to fixation in central Townsville since 2018 (O’Neill et al. 2019) and is now present in several nearby towns (see https://www.worldmosquitoprogram.org/en/global-progress/australia/townsville). Importantly, the invasion may pose a greater biosecurity threat than one from central Townsville, as without the wMel infection, the Tennant Creek Ae. aegypti should be able to transmit dengue without the transmission blocking effects of wMel (Walker et al. 2011). However, our resistance assays show that the invasion remains susceptible to pyrethroid-based control, like other populations in Australia (Endersby-Harshman et al. 2017).
The different patterns of genetic relatedness observed at Tennant Creek and the Torres Strait provide a useful indication of the genetic characteristics of each invasive system, in that we can infer the incursions into the southern Torres Strait Islands are independent from each other, whereas the invasion of Tennant Creek is from a single source. These genetic inferences align with detection locations, where the Tennant Creek samples were collected within ~ 80 m of each other while the Horn and Thursday Island incursions were spread across each island’s range. Understanding the independence of incursions can be useful for building predictive models of pest incursion risk as it can indicate propagule pressure (Camac et al. 2021), though these estimates will reflect effective rather than census population sizes of incursions. The findings at Tennant Creek indicate that greater caution will be required for studies seeking to investigate new invasions using close kin dyad methodologies (Jasper et al. 2022; Waples and Feutry 2022). While close kin dyad methods have shown promise for investigating invasions of ~ 100 generations age (Schmidt et al. 2021c), close kin methods should be applied cautiously to invasions that are very new or that are sourced from a small genetic pool.
Conclusions
The recent and ongoing reduction in sequencing costs has made genomic studies of pests increasingly affordable. A pest genomic databank contains a wealth of information relevant to control, some of which is immediately available (e.g. population structure) and some of which can be revealed over time, such as through tracking the evolution of insecticide resistance or tracing ongoing invasions, incursions, and migration between invaded regions. A concern of pest genomic tracing is that drift and migration will make databank samples obsolete, requiring frequent resampling and sequencing. This study has investigated these concerns, and found that genomic databanks can remain highly powered for precise tracing for at least 70 generations of evolution. Additionally, cross-validation showed that ~ 87% of reference samples could be assigned unambiguously, even though both databanks had low genetic differentiation due to proximity or recent invasion histories. These results suggest that pest genomic databanks can provide valuable tracing insights for many years, even at finer geographical scales. Considering our results specifically, in the Torres Strait, the lack of inbreeding among incursives and the predictability of their origins suggest that the current cordon-sanitaire biosecurity approach (Muzari et al. 2017) is working effectively, as there is no evidence of cryptic establishment on either island. In Tennant Creek, our results highlight the risks of ongoing reestablishment of Ae. aegypti given this invasion was sourced from only a very small number of mosquitoes.
Availability of data and materials
Mosquito .bam files and geographical locations are archived at the NCBI Sequence Read Archive, accessible via accession numbers PRJNA806669 and PRJNA806682.
References
Battey CJ, Ralph PL, Kern AD (2020) Predicting geographic location from genetic variation with deep neural networks. Elife 9:1–22. https://doi.org/10.7554/eLife.54507
Beebe NW, Whelan PI, van den Hurk A et al (2005) Genetic diversity of the dengue vector Aedes aegypti in Australia and implications for future surveillance and mainland incursion monitoring. Commun Dis Intell Q Rep 29:299–304
Beebe NW, Ambrose L, Hill LA et al (2013) Tracing the tiger: Population genetics provides valuable vnsights into the Aedes (Stegomyia) albopictus invasion of the Australasian region. PLOS Negl Trop Dis 7:e2361–e2361. https://doi.org/10.1371/journal.pntd.0002361
Behura SK, Lobo NF, Haas B et al (2011) Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect Biochem Mol 41:770–777. https://doi.org/10.1016/J.IBMB.2011.05.006
Boyle JH, Rastas PMA, Huang X et al (2021) A linkage-based genome assembly for the mosquito Aedes albopictus and identification of chromosomal regions affecting diapause. InSects 12:167. https://doi.org/10.3390/insects12020167
Caley P, Ingram R, De Barro P (2015) Entry of exotic insects into Australia: does border interception count match incursion risk? Biol Invasions 17:1087–1094. https://doi.org/10.1007/s10530-014-0777-z
Chen MZ, Cao LJ, Li BY et al (2021) Migration trajectories of the diamondback moth Plutella xylostella in China inferred from population genomic variation. Pest Manag Sci 77:ps.6188. https://doi.org/10.1002/ps.6188
Danecek P, Auton A, Abecasis G et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Diagne C, Leroy B, Vaissière A-C et al (2021) High and rising economic costs of biological invasions worldwide. Nature 592:571–576. https://doi.org/10.1038/s41586-021-03405-6
Endersby-Harshman NM, Wuliandari JR, Harshman LG et al (2017) Pyrethroid susceptibility has been maintained in the dengue vector, Aedes aegypti (Diptera: Culicidae), in Queensland, Australia. J Med Entomol 54:1649–1658. https://doi.org/10.1093/jme/tjx145
Endersby-Harshman NM, Schmidt TL, Chung J et al (2020) Heterogeneous genetic invasions of three insecticide resistance mutations in Indo-Pacific populations of Aedes aegypti (L.). Mol Ecol 29:1628–1641. https://doi.org/10.1111/mec.15430
Fitzpatrick BM, Fordyce JA, Niemiller ML, Reynolds RG (2012) What can DNA tell us about biological invasions? Biol Invasions 14:245–253. https://doi.org/10.1007/s10530-011-0064-1
Frichot E, François O (2015) LEA : An R package for landscape and ecological association studies. Meth Ecol Evol 6:925–929. https://doi.org/10.1111/2041-210X.12382
Gao Y-F, Gong Y-J, Cao L-J et al (2021) Geographical and interspecific variation in susceptibility of three common thrips species to the insecticide, spinetoram. J Pest Sci 94:93–99. https://doi.org/10.1007/s10340-019-01128-2
Gloria-Soria A, Lima A, Lovin DD et al (2018) Origin of a high-latitude population of Aedes aegypti in Washington, DC. Am J Trop Med Hyg 98:445–452. https://doi.org/10.4269/ajtmh.17-0676
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620. https://doi.org/10.1046/j.1471-8286.2002.00305.x
Hlaing T, Tun-Lin W, Somboon P et al (2009) Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: Implications for past and future population genetic studies. BMC Genet 10:11–11. https://doi.org/10.1186/1471-2156-10-11
Hoffmann A, Turelli M (1997) Cytoplasmic incompatibility in insects. In: O’Neill SL, Hoffmann AA, Werren JH (eds) Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, Oxford, pp 42–80
Hulme PE, Bacher S, Kenis M et al (2008) Grasping at the routes of biological invasions: a framework for integrating pathways into policy. J Appl Ecol 45:403–414. https://doi.org/10.1111/j.1365-2664.2007.01442.x
Jasper ME, Hoffmann AA, Schmidt TL (2022) Estimating dispersal using close kin dyads: The KINDISPERSE R package. Mol Ecol Resour 22:1200–1212. https://doi.org/10.1101/2021.07.28.454079
Kelly ET, Mack LK, Campos M et al (2021) Evidence of local extinction and reintroduction of Aedes aegypti in Exeter, California. Front Trop Dis 2:703873
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lee SF, White VL, Weeks AR et al (2012) High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Appl Environ Microbiol 78:4740–4743. https://doi.org/10.1128/AEM.00069-12
Linck E, Battey CJ (2019) Minor allele frequency thresholds strongly affect population structure inference with genomic data sets. Mol Ecol Resour 19:639–647. https://doi.org/10.1111/1755-0998.12995
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425. https://doi.org/10.1002/j.1537-2197.1995.tb12679.x
Matthews BJ, Dudchenko O, Kingan SB et al (2018) Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 563:501–507. https://doi.org/10.1038/s41586-018-0692-z
Maynard AJ, Ambrose L, Cooper RD et al (2017) Tiger on the prowl: Invasion history and spatio-temporal genetic structure of the Asian tiger mosquito Aedes albopictus (Skuse 1894) in the Indo-Pacific. PLOS Negl Trop Dis 11:e0005546–e0005546. https://doi.org/10.1371/journal.pntd.0005546
Mehta SV, Haight RG, Homans FR et al (2007) Optimal detection and control strategies for invasive species management. Ecol Econ 61:237–245. https://doi.org/10.1016/J.ECOLECON.2006.10.024
Muzari MO, Devine G, Davis J et al (2017) Holding back the tiger: Successful control program protects Australia from Aedes albopictus expansion. PLoS Negl Trop Dis 11:e0005286. https://doi.org/10.1371/journal.pntd.0005286
Myers JH, Simberloff D, Kuris AM, Carey JR (2000) Eradication revisited: dealing with exotic species. Trends Ecol Evol 15:316–320. https://doi.org/10.1016/S0169-5347(00)01914-5
O’Neill SL, Ryan PA, Turley AP et al (2019) Scaled deployment of Wolbachia to protect the community from dengue and other Aedes transmitted arboviruses. Gates Open Res 2:36. https://doi.org/10.12688/gatesopenres.12844.2
Parvizi E, Dhami MK, Yan J, McGaughran A (2023) Population genomic insights into invasion success in a polyphagous agricultural pest, Halyomorpha halys. Mol Ecol 32:138–151. https://doi.org/10.1111/mec.16740
Peterson BK, Weber JN, Kay EH et al (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135–e37135. https://doi.org/10.1371/journal.pone.0037135
Pettit W, Kurucz N (2014) Dengue mosquito eliminated from Tennant Creek again. North Territ Dis Control Bull 21:1–3
Popa-Báez Á-D, Lee SF, Yeap HL et al (2021) Tracing the origins of recent Queensland fruit fly incursions into South Australia, Tasmania and New Zealand. Biol Invasions 23:1117–1130. https://doi.org/10.1007/s10530-020-02422-2
Rašić G, Filipović I, Weeks AR, Hoffmann AA (2014) Genome-wide SNPs lead to strong signals of geographic structure and relatedness patterns in the major arbovirus vector, Aedes aegypti. BMC Genom 15:1–12. https://doi.org/10.1186/1471-2164-15-275
Rašić G, Filipović I, Callahan AG et al (2016) The queenslandensis and the type form of the dengue fever mosquito (Aedes aegypti L.) are genomically indistinguishable. PLOS Negl Trop Dis 10:e0005096. https://doi.org/10.1371/journal.pntd.0005096
Ritchie SA, Moore P, Carruthers M et al (2006) Discovery of a widespread infestation of Aedes albopictus in the Torres Strait, Australia. J Am Mosq Control Assoc 22:358–365. https://doi.org/10.2987/8756-971X(2006)22[358:DOAWIO]2.0.CO;2
Robinson A, Burgman MA, Cannon R (2011) Allocating surveillance resources to reduce ecological invasions: maximizing detections and information about the threat. Ecol Appl 21:1410–1417. https://doi.org/10.1890/10-0195.1
Rochette NC, Rivera-Colón AG, Catchen JM (2019) Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol 28:4737–4754. https://doi.org/10.1111/mec.15253
Ross PA, Wiwatanaratanabutr I, Axford JK et al (2017) Wolbachia infections in Aedes aegypti differ markedly in their response to cyclical heat stress. PLoS Pathog 13:e1006006. https://doi.org/10.1371/journal.ppat.1006006
Ryan PA, Turley AP, Wilson G et al (2020) Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in Cairns and surrounding locations in northern Queensland, Australia. Gates Open Res 3:1547. https://doi.org/10.12688/gatesopenres.13061.2
Sardain A, Sardain E, Leung B (2019) Global forecasts of shipping traffic and biological invasions to 2050. Nat Sustain 2:274–282. https://doi.org/10.1038/s41893-019-0245-y
Schmidt TL, Barton NH, Rašić G et al (2017) Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol 15:e2001894. https://doi.org/10.1371/journal.pbio.2001894
Schmidt TL, Filipović I, Hoffmann AA, Rašić G (2018) Fine-scale landscape genomics helps explain the slow spatial spread of Wolbachia through the Aedes aegypti population in Cairns, Australia. Heredity (edinb) 120:386–395. https://doi.org/10.1038/s41437-017-0039-9
Schmidt TL, van Rooyen AR, Chung J et al (2019) Tracking genetic invasions: genome-wide single nucleotide polymorphisms reveal the source of pyrethroid-resistant Aedes aegypti (yellow fever mosquito) incursions at international ports. Evol Appl 12:1136–1146. https://doi.org/10.1111/eva.12787
Schmidt TL, Chung J, Honnen AC et al (2020a) Population genomics of two invasive mosquitoes (Aedes aegypti and Aedes albopictus) from the Indo-Pacific. PLoS Negl Trop Dis 14:e0008463. https://doi.org/10.1371/journal.pntd.0008463
Schmidt TL, Chung J, van Rooyen AR et al (2020b) Incursion pathways of the Asian tiger mosquito (Aedes albopictus) into Australia contrast sharply with those of the yellow fever mosquito (Aedes aegypti). Pest Manag Sci 76:4202–4209. https://doi.org/10.1002/ps.5977
Schmidt TL, Endersby-Harshman NM, Hoffmann AA (2021a) Improving mosquito control strategies with population genomics. Trends Parasitol 37:907–921. https://doi.org/10.1016/j.pt.2021.05.002
Schmidt TL, Jasper M-E, Weeks AR, Hoffmann AA (2021b) Unbiased population heterozygosity estimates from genome-wide sequence data. Meth Ecol Evol 12:1888–1898. https://doi.org/10.1111/2041-210X.13659
Schmidt TL, Swan T, Chung J et al (2021c) Spatial population genomics of a recent mosquito invasion. Mol Ecol 30:1174–1189. https://doi.org/10.1101/2020.11.30.405191
Sherpa S, Blum MGB, Capblancq T et al (2019) Unravelling the invasion history of the Asian tiger mosquito in Europe. Mol Ecol 28:2360–2377. https://doi.org/10.1111/mec.15071
Stringham OC, Lockwood JL (2021) Managing propagule pressure to prevent invasive species establishments: propagule size, number, and risk–release curve. Ecol Appl 31:e02314. https://doi.org/10.1002/eap.2314
Tobin PC, Berec L, Liebhold AM (2011) Exploiting Allee effects for managing biological invasions. Ecol Lett 14:615–624. https://doi.org/10.1111/j.1461-0248.2011.01614.x
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935. https://doi.org/10.1046/j.1365-294X.2004.02076.x
Walker T, Johnson PH, Moreira LA et al (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453. https://doi.org/10.1038/nature10355
Waples RS, Feutry P (2022) Close-kin methods to estimate census size and effective population size. Fish Fish 23:273–293. https://doi.org/10.1111/faf.12615
Whelan PI, Pettit WJ, Krause VL (2012) Dengue mosquito incursion into Tennant Creek 2011. North Territ Dis Control Bull 19:16–21
Whelan PI, Kurucz N, Pettit WJ, Krause V (2020) Elimination of Aedes aegypti in northern Australia, 2004–2006. J Vector Ecol 45:118–126. https://doi.org/10.1111/jvec.12379
Yan J, Vétek G, Pal C et al (2021) ddRAD sequencing: an emerging technology added to the biosecurity toolbox for tracing the origin of brown marmorated stink bug, Halyomorpha halys (Hemiptera: Pentatomidae). BMC Genom 22:355. https://doi.org/10.1186/s12864-021-07678-z
Yeap HL, Rašić G, Endersby-Harshman NM et al (2016) Mitochondrial DNA variants help monitor the dynamics of Wolbachia invasion into host populations. Heredity (edinb) 116:265–276. https://doi.org/10.1038/hdy.2015.97
Zeigler SL, Fagan WF (2014) Transient windows for connectivity in a changing world. Mov Ecol 2:1. https://doi.org/10.1186/2051-3933-2-1
Camac J, Baumgartner J, Hester S et al (2021) Using edmaps & Zonation to inform multi-pest early-detection surveillance designs. Centre of Excellence for Biosecurity Risk Analysis (CEBRA)
Acknowledgements
We would like to thank all Medical Entomology staff who were involved in mosquito surveillance and identification. We thank Nigel Beebe and Queensland Health for sample collection, and Kelly Richardson for assistance with library construction. We thank two anonymous reviewers for their useful comments.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. Funding was provided by Australian Department of Health, Queensland Health, and the National Health and Medical Research Council (Grant Numbers 1132412, 1118640 and 1131932 (The HOT NORTH initiative)).
Author information
Authors and Affiliations
Contributions
All authors conceived and designed research. NK, WP, VLK, GE, OMM and NEH performed sampling. NEH and TLS generated genetic data. TLS analysed the data. TLS wrote the manuscript, with edits from all authors. All authors approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
We declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schmidt, T.L., Endersby-Harshman, N.M., Kurucz, N. et al. Genomic databanks provide robust assessment of invasive mosquito movement pathways and cryptic establishment. Biol Invasions 25, 3453–3469 (2023). https://doi.org/10.1007/s10530-023-03117-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-023-03117-0