Abstract
The history of human colonisation in the Mediterranean has long been recognised as a crucial factor influencing biodiversity patterns in southern Europe. Nonetheless, our understanding of how anthropogenic and natural dispersal events interacted in shaping wildlife distributions, particularly in small mammals, remains limited. The edible dormouse Glis glis, a widespread European species, whose distribution includes several islands in the Mediterranean, present an opportunity to investigate these interactions. In this work, we used the edible dormouse to test hypotheses regarding the interplay between natural and anthropogenic dispersal in shaping species’ distributions in Mediterranean archipelagos. We compared genetic sequences from samples collected on Mediterranean islands (Elba Island, Corsica, Sardinia, Sicily and Salina Island) and the mainland. Twenty-one samples were analysed by amplifying and sequencing a fragment of the cytochrome oxidase subunit I gene. Results indicated that samples from Sardinia and Elba Island belong to the same clade of mainland Italy, specifically to the subspecies G. g. italicus. This finding does not support the existence of an endemic Sardinian subspecies and suggests recent introduction events. In contrast, Salina Island only included individuals belonging to the Sicilian subspecies, whereas Sicily hosts a mixed population of G. g. italicus and G. g. insularis. The Corsican population likely originated from a different stock than Sardinia, possibly originating from Northern Italy or southern France. Overall, our findings underscore the significant role of anthropogenic dispersal in shaping the current distribution of the edible dormouse on islands.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding connectivity, genetic structure and divergence from neighbouring populations is crucial for planning adequate management actions and conservation strategies (e.g., Garrido-Garduño et al. 2016; Monti et al. 2018), especially for small, isolated and threatened populations (Cresswell 2014). In this context, non-volant land fauna inhabiting islands is an excellent model to assess the potential consequences of insularism, by being particularly prone to reproductive isolation (Barreto et al. 2021). Apart from rats (Rattus spp.) and house mice (Mus musculus / M. domesticus), whose biogeography has long been investigated and influenced by involuntary anthropogenic introductions (e.g., Abdelkrim et al. 2005; Sciandra et al. 2022), small mammals are a relatively untested model to investigate mtDNA genetic polymorphism on islands, which often host endemic lineages for most living species (Yu et al. 1995; Berry 1996; Harr 2006; Nicolas et al. 2014; Iannucci et al. 2019).
During Quaternary climatic oscillations, many northern populations underwent extinction processes, with glacial refugia primary located in southern latitudes, such as in the Mediterranean (Hewitt 2001; Hürner et al. 2010). These processes promoted progressive loss of polymorphism in areas of more recent recolonisation − areas of northward expansions from refugia during interglacial periods − as per the southern refugia hypothesis (Hewitt 2001, 2004; Seddon et al. 2001; but see Pedreschi et al. 2019), and potential founder effects (e.g., Polfus et al. 2017). Nonetheless, the genetic diversity of animal populations from more recently recolonized areas may be even larger than that of populations from southern, previous Last Glacial Maximum refugia (e.g. Markova et al. 2020, Niedziałkowska et al. 2021; Plis et al. 2022). In addition, species may respond to climate oscillations in species-specific ways (Pedreschi et al. 2019) leading to a complex web of subspecific and ecotypic differentiation even within refugia (Ancillotto et al. 2023). Island refugia may thus potentially host populations with specific alleles or genetic structuring, especially for less vagile species with peculiar ecological constraints (e.g., small mammals).
The edible dormouse Glis glis is a small to medium sized rodent (body length 160–190 mm) and, despite narrow ecological preferences associated to forested habitats (Fietz and Weis-Dootz 2012), the species shows a widespread distribution from northern Spain to Russia (Hürner et al. 2010; Krystufek et al. 2021). Paleontological data indicate refugial areas in the Mediterranean region, with low mtDNA gene polymorphism at European level probably due to recent expansion, which likely occurred from a single refugium (Hürner et al. 2010; Michaux et al. 2019). In the past, the species has been intentionally introduced in various locations, including islands (e.g., Masseti 2005) as a food resource and in central-southern England, as a human-mediated unintentional introduction (Burgess et al. 2003).
Traditionally, 21 subspecies of edible dormouse have been identified, although phylogenetic analyses and morphological features appear to support only five of them: (1) G. g. glis, occurring in most of Europe; (2) G. g. minutus, endemic to North Macedonia; (3) G. g. pindicus, confirmed in Alonissos island but possibly also occurring in continental Greece; (4) G. g. italicus, endemic to Central and Southern Italy; (5) G. g. insularis, endemic to Sicily (Hürner et al. 2010; Lo Brutto et al. 2011; Castiglia et al. 2012; Koren et al. 2015; Krystufek et al. 2021). Thus, two clades have been described for islands (namely, G. g. insularis and G. g. pindicus).
Italy hosts the highest number of subspecies of edible dormouse in all Europe. The nominal subspecies is found in northernmost regions, the Italian subspecies is present in central and southern regions, and the Sicilian clade is exclusive to Sicily (Hürner et al. 2010; Lo Brutto et al. 2011; Krystufek et al. 2021). Lo Brutto et al. (2011) claimed that additional sampling from island populations (and/or additional genes) of the species was necessary to clarify their phylogenetic affinities to continental ones. Indeed, the edible dormouse is recorded not only on the major Italian islands (Sicily and Sardinia), but it also occurs in some small islands of the country (e.g., Salina Island on the Aeolian archipelago and Elba Island on the Tuscan Archipelago: Angelici et al. 2009; Hürner et al. 2010), and on the nearby Corsica in France (Sarà 1998). Surprisingly, Amori et al. (2015) found remains of edible dormouse also in a barn owl Tyto alba pellet collected on Capraia Island in the Tuscan Archipelago, although the specimen could have been preyed on the nearby Corsica or Elba Island or on the continent and regurgitated in Capraia at a later time. Similarly, the occurrence of the edible dormouse in Capri (in the Campanian archipelago) reported by Nappi et al. (2007) and by Angelici et al. (2009) relies solely on the record of a local tourist guide (Aprea 1999), and it has never been scientifically confirmed. The distribution of the edible dormouse in Sardinia once included a wider territory and the Asinara island, whereas the species is currently being found only in the main island, where it is locally classified as “Endangered” (Sarà 1998; Hürner et al. 2010; Amori et al. 2014).
In this work, we aimed at clarifying the phylogenetic affinities of the edible dormouse between insular and peninsular populations. We employed a barcoding approach, with particular reference to Elba Island and Corsica, where edible dormice have never been sampled for genetic analyses (e.g., Hürner et al. 2010; Lo Brutto et al. 2011; Michaux et al. 2019). The outcomes of our research will also contribute to a better understanding of the origin of island populations and shed light on possible introduction events that may have occurred in the past.
Materials and methods
We searched for edible dormouse samples by asking for frozen specimens or tissue samples preserved in absolute ethanol to universities (N = 3, from University of Turin), national museums of natural sciences (N = 3 from Museo Civico di Storia Naturale di Carmagnola, N = 2 from Museo di Storia Naturale della Maremma), CNR-IRET collection (N = 8), wildlife recovery centres (N = 2 from CRAS Wild Umbria) and local environmental organizations (N = 1 from Associazione Nesos) (Table 1). All samples we included in our work were labelled with unequivocal names and geographic location origins.
We focused on the cytochrome oxidase I gene of the mitochondrial DNA (hereafter, COXI), as other genes have already been extensively studied for this species (e.g., the cytochrome-b and the D-loop), and authors of previous studies have emphasized for the importance of considering other genes to better define the phylogenetic position of insular populations of Italy (e.g., see Hürner et al. 2010; Lo Brutto et al. 2011).
All samples were collected between 2012 and 2023 and were either stored in absolute ethanol or frozen.
We extracted total genomic DNA from 25 mg of dormouse tissue using the Qiagen Blood and Tissue kit (©Qiagen, Inc, Tokyo, Japan). We amplified a portion (638 bp) of the COXI by using universal primers: HCO 2198 (5’-TAAACTTCAGGGTGACCAAAAAATCA-3’) and LCO 1490 (5’- GGTCAACAAATCATAAAGATATTGG-3’) (Folmer et al. 1994). We conducted the PCR reactions on an Eppendorf MasterCycler X50 thermal cycler in 25 µL mix including 100 ng of each DNA sample, buffer 10×, 1.2 mM MgCl2, 200 µM dNTPs, 0.2 µM of each primer, and one unit of Taq polymerase (©Life Technologies, Waltham, Massachusetts, USA). PCR conditions included initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 45′′, annealing at 50 °C for 30′′, extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min (Mori et al. 2022). PCR products were run by electrophoresis on 2.0% agarose gels containing 0.5 mg/mL of SYBR gel staining. Successful amplifications were then purified (ExoSAP-IT PCR clean-up Kit, ©Applied Biosystems, Foster City, California, USA) and sequenced via the chain termination method at the BMR Genomics (Padova, Italy). Obtained sequences were corrected and aligned with the Mega XI software (Tamura et al. 2021). Nucleotide diversity, haplotype diversity and number of polymorphic sites were computed through DNAsp vers. 5 (Librado and Rozas 2009). The software JModelTEST 304 (Posada and Crandall 1998) was used to test the most accurate model of substitution using the Bayesian Information Criterion (BIC) and Akaike’s Information Criterion (AIC), corrected for the heterogeneity among sites (gamma [G]). Dormouse sequences obtained in this study were aligned with previously published COXI sequences of the same species available on GenBank (http://www.ncbi.nlm.nih.gov): one from Austria (accession number: KY754509), two from France (accession numbers: AJ001562 and NC001892), three from Switzerland (accession numbers: MW478021, OQ706750 and OQ706751), and four from Poland (accession numbers: FJ808614, FJ808615, FJ808616 and FJ808617). The Tamura-Nei 93 nucleotide substitution model was selected. We conducted phylogenetic reconstruction applying maximum likelihood (with the software SeaView: Gouy et al. 2010), using the best model selected. Four chains of Markov Chain Monte Carlo were run simultaneously and sampled every 1000 generations for 4 million generations. The first 1000 sampled trees from each run were discarded as burn-in. We selected optimized choices, and we obtained the tree-searching operations by Nearest-Neighbour Interchange (NNI) and Subtree Pruning–Regrafting (SPR). A sequence of forest dormouse Dryomys nitedula from northern Italy (46°41’27.6"N 11°19’44.4"E: accession number: MZ661159) was included as an outgroup.
A TCS network (Clement et al. 2000) connecting haplotypes was obtained with the software popART (Available at https://popart.otago.ac.nz, accessed on 10.10.2023) to visualise the relationship between the new and the previously described haplotypes of the edible dormouse (Sciandra et al. 2022).
A Principal Coordinate Analysis (PCoA) was used to assess the genetic structuring of the studied species through the software GenAlEx 6 (Peakall and Smouse 2006). We estimated the occurrence of molecular operational taxonomic unit (MOTU) to infer species delimitation criteria based on a partial COXI gene by employing the Automatic Barcode Gap Discovery (ABGD: Puillandre et al. 2012), which was run on the ABGD web server (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html: accessed on 07.04.2024). The ABGD separates the species based on a range of maximum intraspecific distance (set parameters: steps = 10, X = 1.5).
Results
We were able to collect 21 samples of edible dormouse (1–3 per location, amplicon size = 638 bp, N = 48 polymorphic sites, nucleotide diversity p = 0.026, haplotype diversity h = 0.759: cf. Table 1), two of which from Elba Island (province of Livorno, Tuscany), two from Salina Island (Aeolian archipelago, Sicily), one from Sardinia (Urzulei, Nuoro), one from Corsica (Vizzavona), and one from Sicily (Ficuzza, Palermo). All the other samples came from peninsular Italy (Table 1). Genetic distances among different subspecies are shown in Table 2.
Samples from Elba Island and Sardinia clustered with peninsular samples from Central Italy, while a second supported cluster was composed by Salina Island and Sicilian samples, consistently with the described subspecies G. g. italicus and G. g. insularis (Figs. 1 and 2). Samples from Northern Italy, including both the Alps and the northern Apennines, clustered with a sample from Corsica in a supported subcluster of the G. g. glis subspecies, which also include sequences from Poland, Switzerland, Austria and continental France (Fig. 1).
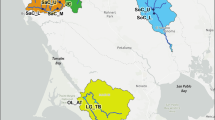
Right, Italian map of edible dormouse sampes: G. g. insularis is represented by amber triangles, G. g. glis by yellow dots and G. g. italicus by green squares. European map in the inset shows the origin of all samples used in this study. Left, maximum likelihood phylogenetic tree of Glis glis using COXI sequences (638 bp). Bootstrap values higher than 90% are indicated at tree nodes
TCS network showing the relationship among new and previously described haplotypes of edible dormouse. Circles represent different haplotypes. The green circle represents the haplotype from all localities of Central and Southern Italy, including samples from Sardinia and Elba Island. The amber circle represents the haplotype from Sicily, including the samples from Salina Island, while yellow circles represent the nominal subspecies including samples from Northern Italy, Austria, Switzerland, Continental France, Corsica and Poland. Circle sizes are proportional to the number of specimens examined for each haplotype, while dashes indicate mutational steps
We recorded a polytomy within the cluster corresponding to the italicus group, which may suggest a lack of internal differentiation. The italicus and insularis groups represented sister clusters, as confirmed also by the reduced genetic distances with respect to the nominal subspecies (Table 2). As to the nominal subspecies, two sister clusters were supported, representing continental Europe and the Italian peninsula (Fig. 1).
The TCS network (Fig. 2) and the PCoA (Fig. 3) also showed a structuration of Italian edible dormouse in three haplogroups, with the southern Sicilian one being more differentiated from the others, according to the higher number of mutational steps (n = 21).
Only the structuring along the x-axis of PCoA, which explains 43% of the variability, is sufficiently supported: insularis and italicus groups are separated in the multivariate space along the y-axis which explained only 23% of the variability.
Particularly, all samples from peninsular Italy, Sardinia and Elba Island clustered in the same haplotype. Indeed, eight haplotypes were identified for samples of the nominal subspecies, one of which including all those coming from Northern Italy and Corsica, whereas a single haplotype included all sequences of G. g. italicus and another one those of G. g. insularis (Fig. 2).
Results obtained by ABGD suggested the occurrence of three taxonomic units, i.e. the same detected by the phylogenetic analyses, given their limited distance (Fig. 4).
Discussion
In our work, we presented the first molecular phylogeny using the COXI gene to analyse all Italian and Western Mediterranean Island populations of edible dormice. Specifically, samples were collected on all islands where this species is known to occur, i.e., Sardinia, Sicily, Elba Island, Salina Island and Corsica. The reported presence of this species in Asinara, Capri and Capraia islands remains unconfirmed and may be attributed to past misidentifications (e.g., with the garden dormouse Eliomys quercinus in Asinara: Amori et al. 2009), references to archaeological data (e.g., in Capri: Masseti 2005) or individual occurrences found in owl pellets, likely predated upon in other areas (for Capraia island: Amori et al. 2015).
Our limited sample size was due to the small population size of the edible dormouse on Italian island and Corsica, and to the consequent scarcity of specimens in museum collections (Amori et al. 2009). Nevertheless, our analyses confirmed the presence of three divergent populations, one occurring in Northern regions, one in Peninsular regions and Sardinia and the third one in Sicily.
Our findings align with the genetic lineages identified across Europe by Hürner et al. (2010), confirming the established subspecific classification. The northern Italian haplotype belongs to the nominal subspecies found throughout Europe, although it constitutes a separate and divergent sub-clade with respect to the rest of European samples, which also includes the Corsican sample.
In Central Italy, including Sardinia, the only haplotype present is the G. g. italicus. Moving southwards to Sicily and Salina Island, a third distinct haplotype, i.e. G. g. insularis, is observed (Lo Brutto et al. 2011), representing the sister taxon of G. g. italicus. These three haplotypes exhibit well-defined and non-overlapping geographical distribution from north to south. However, this low genetic variability of the COXI for peninsular Italy (G. g. italicus) and Sicily (G. g. insularis) might be due to a founder effect. It is worth noting that Hürner et al. (2010) found a partial overlap in Sicily between G. g. italicus and G. g. insularis. Our PCoA also supports the low genetic difference between insularis and italicus groups, further confirming the genetic affinity between these groups evidenced by the phylogenetic tree. This result is phylogenetically reasonable as it suggests that there may have been some gene flow from mainland Italy to Sicily, where a different taxonomic unit has recently differentiated (Amori et al. 2008). Moreover, as observed for several other species (e.g., Amori and Castiglia 2018; Ancillotto et al. 2023), the nominal subspecies of the edible dormouse in Italy formed a divergent subgroup with respect to samples from the rest of Europe, consistently with the role of glacial refugium of the Italian peninsula. Further genes (e.g., 16 S–12 S) should be amplified to reliably estimate the temporal origin of the three groups (Rocha-Olivares et al. 2001; Gaunt and Miles 2002).
Previous phylogenetic studies on the species have indicated an extremely limited genetic polymorphism, potentially rendering the species highly vulnerable to future climate changes that could pose a threat to its long-term conservation (e.g., Hürner et al. 2010). In this context, Italy assumes a pivotal role for the conservation and evolutionary potential of the edible dormouse, as it hosts the greatest diversity of haplogroups at the European level.
The genetic distance of the haplotype of southern group represented by Sicilian samples aligns with the recognised role played by southern regions and islands as glacial refugia. This observation is consistent with the earlier results reported by Hürner et al. (2010) and is supported by fossil records (Fiore et al. 2004). Palaeontological evidence suggests that the dispersal of the representatives of the Gliridae family likely occurred only on the islands of the western part of the Mediterranean basin, during the Upper Pleistocene (Masseti 2005).
Regarding specimens from the newly sampled localities, Elba Island and Corsica, our findings revealed that, despite their geographical proximity and limited distance, such as with Sardinia, they belong to two different clusters. Samples from Elba Island cluster with the italicus clade, whereas the Corsican sample seems to belong to the nominal subspecies distributed across the rest of Europe. The Corsican sample exhibits greater similarity to specimens from Northern Italy, particularly Piedmont, than to neighbouring samples from Central Italy and to continental France ones. This dissimilarity may be indicative of distinct introduction events on the two islands in historical times. In the case of Corsica, introduction likely occurred with individuals originally coming from Northern Italy, and it may have occurred in recent times, approximately 100–150 years ago (Major 1905), although some historical records also date back to 4000–5000 years ago (Vigne 1992). However, additional samples are necessary to confirm this hypothesis.
The first known record of edible dormouse on Elba Island dates back to the early XIX Century (Thiebaut de Bernaud 1808; Angelici et al. 2009). Nevertheless, Caloi et al. (1986) also reported that during the final stages of the last Ice Age, many islands close to the coast, e.g. the Elba Island, remained connected to the mainland for an extended period. This connectivity would have facilitated the spread of mainland species to these islands, which may explain the observed genetic similarity. However, the local lack of fossil record militates from an introduction from coastal Central Italy.
The current distribution of the edible dormouse results from both evolutionary differentiation processes over time and human-mediated introduction events in ancient times. Historically, the edible dormouse was commonly imported, and kept in large walled enclosures known as “gliraria” for fattening before being consumed as a delicacy (Carpaneto and Cristaldi 1995; Masseti 2005; Meléndez and Campos Carrasco 2023). This practice likely facilitated the translocation of individuals to various locations, including Sardinia and Salina Island (Cristaldi and Amori 1982; Angelici et al. 2009), and the emergence of populations with distinctive genetic structure in different areas. The introduction process may have involved individuals from different regions, potentially explaining the admixture of clades, such as G. g. italicus and G. g. insularis observed in Sicily (Hürner et al. 2010). Despite the limited sample size for these locations, these results contribute to a more precisely delineation of the distribution limit of the two subspecies between Central and Northern Italy, including the islands. This unveils a phylogeographic pattern consistent with both natural and anthropogenic evolutionary processes, as typical with small vertebrates from Mediterranean islands (Ancillotto et al. 2020; Fichera et al. 2022; Sciandra et al. 2022; Antinucci et al. 2023). In most cases, recent or ancient introductions of edible dormice on island have failed. However, in some instances (Elba, Sardinia, Corsica) current populations of this rodent are genetically very similar to those occurring on the mainland, indicating the success of introductions. Therefore, if confirmed also through nuclear genes, the identification of the endemic Sardinian subspecies G. g. meloni might be regarded as a junior synonym to G. g. italicus (Kryštufek et al. 2021). Conversely, differently from Sicily, Salina Island seems to host only individuals of the southern subspecies G. g. insularis, which may deserve special conservation measures.
Humans have been, and continue to be, responsible for altering natural dispersal patterns and gene flow among populations in multiple ways (Crispo et al. 2011). Human-mediated reshuffling in species distribution ranges and genetic integrity are frequently unexpected and challenging to monitor, underscoring the importance of comprehensive surveillance, integrative particularly for understudied or neglected taxa such as small rodents.
References
Abdelkrim J, Pascal M, Samadi S (2005) Island colonization and founder effects: the invasion of the Guadeloupe islands by ship rats (Rattus rattus). Molec Ecol 14:2923–2931
Amori G, Castiglia R (2018) Mammal endemism in Italy: a review. Biogeogr 33:19–31
Amori G, Contoli L, Nappi A (2009) Mammalia II. Erinaceomorpha, Soricomorpha, Lagomorpha, Rodentia. Fauna d’Italia. Edizioni Calderini, Bologna, Italy
Amori G, Luiselli L, Milana G, Casula M (2014) Distribuzione, diversità e abbondanza di micromammiferi associati ad habitat forestali in Sardegna. Technical Report. Regione Autonoma della Sardegna and CNR-ISE Editions, Ente Foreste della Sardegna, Cagliari, Italy
Amori G, Rizzo Pinna V, Sammuri G, Luiselli L (2015) Diversity of small mammal communities of the Tuscan Archipelago: testing the effects of island size, distance from mainland and human density. Folia Zool 64:161–166
Ancillotto L, Bosso L, Smeraldo S, Mori E, Mazza G, Herkt M, Galimberti A, Ramazzotti F, Russo D (2020) An African bat in Europe, Plecotus gaisleri: Biogeographic and ecological insights from molecular taxonomy and species distribution models. Ecol Evol 10(12):5785–5800
Ancillotto L, Viviano A, Baratti M, Sogliani D, Ladurner E, Mori E (2023) Every branch in its niche: intraspecific variation in habitat suitability of a widely distributed small mammal, the harvest mouse Micromys minutus. Mammal Research, pp 1–11
Angelici FM, Laurenti A, Nappi A (2009) A checklist of the mammals of small Italian islands. Hystrix 20:3–27
Antinucci C, Gallozzi F, Ancillotto L, Mori E, Castiglia R (2023) Lizards on the borders: source and patterns of colonization of an opportunistic reptile, Podarcis Siculus, on the remote island of Pantelleria (Italy) depicted by mtDNA phylogeography and dorsal pattern. Biologia 78(12):3479–3485
Aprea G (1999) Guida naturalistica all’Isola di Capri. Edizioni La Conchiglia, Capri: 134 pp
Barreto E, Rangel TF, Pellissier L, Graham CH (2021) Area, isolation and climate explain the diversity of mammals on islands worldwide. Proc R Soc B 288:20211879
Berry RJ (1996) Small mammal differentiation on islands. Phil Trans R Soc Lond Ser B: Biol Sci 351:753–764
Burgess M, Morris P, Bright P (2003) Population dynamics of the edible dormouse (Glis glis) in England. Acta Zool Ac Sci Hung 49:27–31
Caloi L, Gliozzi E, Kotsakis T, Malatesta A, Palombo MR (1986) On the paleobiogeography of pleistocenic Italian mammals. Hystrix 1:7–23
Carpaneto G, Cristaldi M (1995) Dormice and man: a review of past and present relations. Hystrix 6:303–330
Castiglia R, Annesi F, Cattaneo C, Grano M, Milana G, Amori G (2012) A new mitochondrial lineage in the edible dormouse, Glis glis (Rodentia: Gliridae), from Alonissos island (Sporades Archipelago, Greece). Folia Zool 61:177–180
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Molec Ecol 9:1657–1660
Cresswell W (2014) Migratory connectivity of Palaearctic African migratory birds and their responses to environmental change: the serial residency hypothesis. Ibis 156:493–510
Crispo E, Moore JS, Lee-Yaw JA, Gray SM, Haller BC (2011) Broken barriers: human‐induced changes to gene flow and introgression in animals: an examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays 33(7):508–518
Cristaldi M, Amori G, de Messine (1982) Italie) Mammalia 46: 405–407
de Thiebaut A (1808) Voyage à l’île d’Elbe, suivi d’une notice sur les autres îles de la Mer Tyrrhenienne. D. Colas & Le Normant, Paris, p 234
Fichera G, Mucedda M, Russo D, Tomassini A, Kiefer A, Veith M, Ancillotto L (2022) Pantelleria island (Sicily, Italy): a biogeographic crossroad for bats between Africa and Europe. Hystrix Italian J Mammalogy 33(2):134–137
Fietz J, Weis-Dootz T (2012) Stranded on an island: consequences of forest fragmentation for body size variations in an arboreal mammal, the edible dormouse (Glis glis). Popul Ecol 54:313–320
Fiore I, Pino Uria B, Tagliacozzo A (2004) L’exploitation Des Petits Animaux Au Paléolithique supérieur–Mésolithique en italie: L’exemple De La Grotta Del Santuario della Madonna De Praia a mare (Cosenza, Italie). Petits Animaux et sociétés humaines: du complément alimentaire aux ressources utilitaires. Actes des rencontres 23–25 octobre 2003. APDCA, Antibes, France, pp 417–430
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molec Mar Biol Biotechn 3:294
Garrido-Garduño T, Téllez‐Valdés O, Manel S, Vázquez‐Domínguez E (2016) Role of habitat heterogeneity and landscape connectivity in shaping gene flow and spatial population structure of a dominant rodent species in a tropical dry forest. J Zool 298:293–302
Gaunt MW, Miles MA (2002) An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol 19:748–761
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molec Biol Evol 27:221–224
Harr B (2006) Genomic islands of differentiation between house mouse subspecies. Genome Res 16:730–737
Hewitt GM (2001) Speciation, hybrid zones and phylogeography- Or seeing genes in space and time. Molec Ecol 10:537–549
Hewitt GM (2004) The structure of biodiversity–insights from molecular phylogeography. Front Zool 1:4–16
Hürner H, Krystufek B, Sarà M, Ribas A, Ruch T, Sommer R, Ivashkina V, Michaux JR (2010) Mitochondrial phylogeography of the edible dormouse (Glis glis) in the western Palearctic region. J Mammal 91:233–242
Iannucci A, Baccetti N, Giannini F, Gotti C, Baratti M (2019) A genetic analysis of the European hedgehog (Erinaceus europaeus): an applicative case study to support its eradication from Pianosa Island (Tuscan Archipelago). Conserv Gen 20:395–402
Koren T, Jelić M, Kryštufek B (2015) Mitochondrial sequences yield new insight into the quaternary history of the edible dormouse on the landbridge Adriatic islands. Mammal Biol 80:128–134
Kryštufek B, Naderi M, Janžekovič F, Hutterer R, Bombek D, Mahmoudi A (2021) A taxonomic revision of fat dormice, genus Glis (Rodentia). Mammalia 85:362–378
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lo Brutto S, Sarà M, Arculeo M (2011) Italian Peninsula preserves an evolutionary lineage of the fat dormouse Glis glis L. (Rodentia: Gliridae). Biol J Linn Soc 102:11–21
Major CF (1905) Rodents from the Pleistocene of the Western Mediterranean Region. Geol Mag 2:462–467
Marková S, Horníková M, Lanier HC, Henttonen H, Searle JB, Weider LJ, Kotlík P (2020) High genomic diversity in the bank Vole at the northern apex of a range expansion: the role of multiple colonizations and end-glacial refugia. Mol Ecol 29:1730–1744
Masseti M (2005) Natural and anthropochorous squirrels and dormice of the Mediterranean region. Hystrix 16:3–26
Meléndez JB, Campos Carrasco JM (2023) Vivaria in Doliis. Ceramic jars for dormouse fattening found in Arucci. Oxf J Archaeol 42:244–255
Michaux JR, Hürner H, Krystufek B, Sarà M, Ribas A, Ruch T, Vekhnik V, Renaud S (2019) Genetic structure of a European forest species, the edible dormouse (Glis glis): a consequence of past anthropogenic forest fragmentation? Biol J Linn Soc 126:836–851
Monti F, Delfour F, Arnal V, Zenboudji S, Duriez O, Montgelard C (2018) Genetic connectivity among osprey populations and consequences for conservation: philopatry versus dispersal as key factors. Conserv Genet 19:839–851
Mori E, Viviano A, Mazzotti S, Sogliani D, Bini A, Baratti M (2022) Unveiling the genetic diversity of declining population of the harvest mouse Micromys minutus in Italy. Diversity 14:627
Nappi A, Cipolla RM, Gabriele R, Masseti M, Corti C, Arcidiacono G (2007) Anfibi, Rettili E Mammiferi delle isole del golfo di Napoli: check-list commentata. Studi Trentini Sci Nat Acta Biologica 83:93–97
Nicolas V, Hamani A, Amrouche L, Bensidhoum M, Boukhemza M, Doumandji S, Denys C (2014) First molecular evidence for the presence of Crocidura pachyura (Mammalia, Soricidae) in Kabylie (Algeria). Mammalia 78:245–249
Niedziałkowska M, Tarnowska E, Ligmanowska J, Jędrzejewska B, Podgórski T, Radziszewska A, Ratajczyk I, Kusza S, Bunevich AN, Danila G, Shkvyria M, Grzybowski G, Woźniak M (2021) Clear phylogeographic pattern and genetic structure of wild boar Sus scrofa population in Central and Eastern Europe. Sci Rep 11:9680
Peakall ROD, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pedreschi D, García-Rodríguez O, Yannic G, Cantarello E, Diaz A, Golicher D, Korstjens AG, Heckel G, Searle JB, Gillingham P, Hardouin EA, Stewart JR (2019) Challenging the European southern refugium hypothesis: species‐specific structures versus general patterns of genetic diversity and differentiation among small mammals. Global Ecol Biogeogr 28:262–274
Plis K, Niedziałkowska M, Borowik T, Lang J, Heddergott M, Tiainen J, Bunevich A, Šprem N, Paule L, Danilkin A, Kholodova M, Zvychaynaya E, Kashinina N, Pokorny B, Flajšman K, Paulauskas A, Djan M, Ristić Z, Novák L, Kusza S, Miller C, Tsaparis D, Stoyanov S, Shkvyria M, Suchentrunk F, Kutal M, Lavadinović V, Šnjegota D, Krapal AM, Dănilă G, Veeroja R, Dulko E, Jędrzejewska B (2022) Pan-european phylogeography of the European roe deer (Capreolus capreolus). Ecol Evol 12:e8931
Polfus JL, Manseau M, Klütsch CF, Simmons D, Wilson PJ (2017) Ancient diversification in glacial refugia leads to intraspecific diversity in a holarctic mammal. J Biogeogr 44:386–396
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species Delimitation. Mol Ecol 21:1864–1877
Reyes A, Pesole G, Saccone C (1998) Complete mitochondrial DNA sequence of the fat dormouse, Glis glis:further evidence of rodent paraphyly. Mol Biol Evol 15:499–505
Rocha-Olivares A, Fleeger JW, Foltz DW (2001) Decoupling of molecular and morphological evolution in deep lineages of a meiobenthic harpacticoid copepod. Mol Biol Evol 18:1088–1102
Ruedi M, Manzinalli J, Dietrich A, Vinciguerra L (2023) Shortcomings of DNA barcodes: a perspective from the Mammal fauna of Switzerland. Hystrix 34:54–61
Sarà M (1998) I mammiferi delle isole del Mediterraneo. L’Epos, Palermo, Italy
Schäffer S, Zachos FE, Koblmüller S (2017) Opening the treasure chest: a DNA-barcoding primer set for most higher taxa of central European birds and mammals from museum collections. PLoS ONE 12:e0174449
Sciandra C, Mori E, Solano E, Mazza G, Viviano A, Scarfò M, Bona F, Annesi F, Castiglia R (2022) Mice on the borders: genetic identification of rat and house mouse species in Lampedusa and Pantelleria islands (Southern Italy). Biogeogr 37:a013
Seddon JM, Santucci F, Reeve NJ, Hewitt GM (2001) DNA footprints of European hedgehogs, Erinaceus europaeus and E. Concolor: pleistocene refugia, postglacial expansion and colonization routes. Mol Ecol 10:2187–2198
Tamura K, Stecher G, Kumar S (2021) Mega11: molecular evolutionary genetics analysis version 11. Molec Biol Evol 38:3022–3027
Vigne JD (1992) Zooarchaeology and the biogeographical history of the mammals of Corsica and Sardinia since the last ice age. Mammal Rev 22:87–96
Yu HT (1995) Patterns of diversification and genetic population structure of small mammals in Taiwan. Biol J Linn Soc 55:69–89
Acknowledgements
Authors are deeply indebted with Dr. Mariella Baratti (CNR-IRET) for her qualified lab and bioinformatic work, who helped throughout the molecular analyses. Authors would like to thank Leonardo Brustenga and Ilaria Guy, who provided us with reference samples from the Italian peninsula. Pietro Lo Cascio, from Associazione Nesos, who provided one sample from Salina Island. DS is grateful to the Ghislieri Foundation for supporting his research activity with an accommodation scholarship. Two anonymous reviewers and the AE kindly improved our MS with their comments.
Funding
EM and LA were funded by the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4 - Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, CUP B83C22002930006, Project title “National Biodiversity Future Center - NBFC”. FM was funded by EU - Next Generation EU Mission 4 “Education and Research” - Component 2: “From research to business” - Investment 3.1: “Fund for the realization of an integrated system of research and innovation infrastructures” - Project IR0000032 – ITINERIS - Italian Integrated Environmental Research Infrastructures System - CUP B53C22002150006. The authors acknowledge the Research Infrastructures participating in the ITINERIS project with their Italian nodes: ACTRIS, ANAEE, ATLaS, CeTRA, DANUBIUS, DISSCO, e-LTER, ECORD, EMPHASIS, EMSO, EUFAR, Euro-Argo, EuroFleets, Geoscience, IBISBA, ICOS, JERICO, LIFEWATCH, LNS, N/R Laura Bassi, SIOS, SMINO.
Open access funding provided by Consiglio Nazionale Delle Ricerche (CNR) within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
EM, FM and LA conceived this work and planned the analyses; DS, FV, SB, GA and GB collected tissue samples; AV, LA and EM conducted genetic analyses and bioinformatic elaborations. EM, DS and FM wrote the first draft; all authors participated in writing the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Authors certify that no living animal was used for this research. All genetic sequences are deposited on GenBank and all samples are available for further analyses.
Conflict of interest
Authors certify that they have no affiliation with or involvement in any organization or entity with any financial or nonfinancial interest in the subject matter or materials discussed in this manuscript. Therefore, they have no conflict of interest to declare.
Additional information
Communicated by Magdalena Niedziałkowska.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mori, E., Ancillotto, L., Viviano, A. et al. Anthropogenic dispersal explains the phylogeography of insular edible dormouse Glis glis in the Mediterranean basin. Mamm Res (2024). https://doi.org/10.1007/s13364-024-00754-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13364-024-00754-1