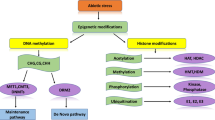
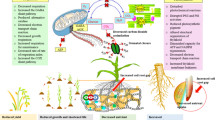
Abstract
Environmental factors, such as drought, salinity, extreme temperature, ozone poisoning, metal toxicity etc., significantly affect crops. To study these factors and to design a possible remedy, biological experimental data concerning these crops requires the quantification of gene expression and comparative analyses at high throughput level. Development of microarrays is the platform to study the differential expression profiling of the targeted genes. This technology can be applied to gene expression studies, ranging from individual genes to whole genome level. It is now possible to perform the quantification of the differential expression of genes on a glass slide in a single experiment. This review documents recently published reports on the use of microarrays for the identification of genes in different plant species playing their role in different cellular networks under abiotic stresses. The regulation pattern of differentially-expressed genes, individually or in group form, may help us to study different pathways and functions at the cellular and molecular level. These studies can provide us with a lot of useful information to unravel the mystery of abiotic stresses in important crop plants.


Similar content being viewed by others
References
Aoki K, Yano K, Suzuki A et al (2010) Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics. BMC Genom 11:210
Bita CE, Zenoni S, Vriezen WH et al (2011) Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genom 12:384
Brandl J, Anderson MR (2015) Current state of genome-scale modeling in filamentous fungi. Biotechnol Lett 37:1131–1139
Cai G-L, Wang X-J, Liu D et al (2010) Isolation and expression analysis of a TaOZR gene induced by stripe rust fungus in wheat. Sci Agric Sin 12:001
Cai H, Tian S, Liu C et al (2011) Identification of a MYB3R gene involved in drought, salt and cold stress in wheat (Triticum aestivum L.). Gene 485:146–152
Cai H, Lu Y, Xie W et al (2012) Transcriptome response to nitrogen starvation in rice. J Biosci 37:731–747
Chang-Kug K, Lim H-M, Na J-K et al (2014) A multistep screening method to identify genes using evolutionary transcriptome of plants. Evol Bioinform Online 10:69
Chen Y, Zong J, Tan Z et al (2015) Systematic mining of salt-tolerant genes in halophyte-Zoysia matrella through cDNA expression library screening. Plant Physiol Biochem 89:44–52
Chmielowska-Bąk J, Lefèvre I, Lutts S et al (2013) Short term signaling responses in roots of young soybean seedlings exposed to cadmium stress. J Plant Physiol 170:1585–1594
Coram TE, Huang X, Zhan G et al (2010) Meta-analysis of transcripts associated with race-specific resistance to stripe rust in wheat demonstrates common induction of blue copper-binding protein, heat-stress transcription factor, pathogen-induced WIR1A protein, and ent-kaurene synthase transcripts. Funct Integr Genom 10:383–392
DalCorso G, Farinati S, Furini A (2010) Regulatory networks of cadmium stress in plants. Plant Signal Behav 5:663–667
D’Andrea RM, Triassi A, Casas MI et al (2015) Identification of genes involved in the drought adaptation and recovery in Portulaca oleracea by differential display. Plant Physiol Biochem 90:38–49
Das R, Pandey GK (2010) Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr Genom 11:2
Duan J, Tian X, Jia Z (2013) Proteomics uncovers a role for enhanced ultraviolet-B radiation on wheat leaves. Amer J Plant Sci 4:1227–1232
Frei M, Tanaka JP, Chen CP et al (2010) Mechanisms of ozone tolerance in rice: characterization of two QTLs affecting leaf bronzing by gene expression profiling and biochemical analyses. J Exp Bot 61:1405–1417
Gong P, Zhang J, Li H et al (2010) Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J Exp Bot 61:3563–3575
Gu L, Liu Y, Zong X et al (2010) Overexpression of maize mitogen-activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol Biol Rep 37:4067–4073
Guo M, Rupe MA, Wei J et al (2014) Maize ARGOS1 (ZAR1) transgenic alleles increase hybrid maize yield. J Exp Bot 65:249–260
Hacham Y, Koussevitzky S, Kirma M et al (2014) Glutathione application affects the transcript profile of genes in Arabidopsis seedling. J Plant Physiol 171:1444–1451
Han YY, Li AX, Li F et al (2012) Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol Biochem 54:49–58
Hasthanasombut S, Paisarnwipatpong N, Triwitayakorn K et al (2011) Expression of OsBADH1 gene in Indica rice (Oryza sativa L.) in correlation with salt, plasmolysis, temperature and light stresses. Plant Omics 4:75–81
Hossain Z, Khatoon A, Komatsu S (2013) Soybean proteomics for unraveling abiotic stress response mechanism. J Proteom Res 12:4670–4684
Huertas R, Rubio L, Cagnac O et al (2013) The K +/H + antiporter LeNHX2 increases salt tolerance by improving K + homeostasis in transgenic tomato. Plant Cell Environ 36:2135–2149
Humbert S, Subedi S, Cohn J et al (2013) Genome-wide expression profiling of maize in response to individual and combined water and nitrogen stresses. BMC Genom 14:3
Jain D, Chattopadhyay D (2010) Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol 10:24
Janská A, Maršík P, Zelenková S et al (2010) Cold stress and acclimation–what is important for metabolic adjustment? Plant Biol 12:395–405
Jia-Shing C, Lin SC, Chen CY et al (2014) Development of microarray for two rice subspecies: characterization and validation of gene expression in rice. BMC Res Note 7:15
Karakach TK, Flight RM, Douglas SE et al (2010) An introduction to DNA microarrays for gene expression analysis. Chemom Intel Lab Sys 104:28–52
Kong X, Li D (2011) Hydrogen peroxide is not involved in HrpN from Erwinia amylovora-induced hypersensitive cell death in maize leaves. Plant Cell Rep 30:1273–1279
Kong X, Sun L, Zhou Y et al (2011) ZmMKK4 regulates osmotic stress through reactive oxygen species scavenging in transgenic tobacco. Plant Cell Rep 30:2097–2104
Kreslavski V, Los D et al (2012) Signalling role of reactive oxygen species in plants under stress. Russ J Plant Physiol 59:141–154
Lee S-H, Lee K-W, Lee D-G et al (2015) Identification and functional characterization of Siberian wild rye (Elymus sibiricus L.) small heat shock protein 16.9 gene (EsHsp16.9) conferring diverse stress tolerance in prokaryotic cells. Biotechnol Lett 37:881–890
Li F, Xing S, Guo Q et al (2011) Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J Plant Physiol 168(9):960–966
Li C-Y, Deng G-M, Yang J et al (2012) Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom 13:374
Li C, Yan JM, Li YZ et al (2013) Silencing the SpMPK1, SpMPK2, and SpMPK3 genes in tomato reduces abscisic acid-mediated drought tolerance. Int J Mol Sci 14:21983–21996
Li Z, Wang Z, Li S (2015) Gene chip analysis of Arabidopsis thaliana genomic DNA methylation and gene expression in response to carbendazim. Biotechnol Lett 37:1297–1307
Liu H, Yang W, Liu D et al (2011) Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Mol Biol Rep 38:417–427
Liu G-T, Wang J-F, Cramer G et al (2012) Transcriptomic analysis of grape (Vitis vinifera L.) leaves during and after recovery from heat stress. BMC Plant Biol 12:174
Luo M, Liu J, Lee RD et al (2010) Monitoring the expression of maize genes in developing kernels under drought stress using oligomicroarray. J Integ Plant Biol 52:1059–1074
Ma T-L, Wu W-H, Wang Y (2012) Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12:161
Mao X, Zhang H, Qian X et al (2012) TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 63:2933–2946
Mariutto M, Duby F, Adam A et al (2011) The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol 11:29
Meng J, Li R, Luan Y (2015) Classification by integrating plant stress response gene expression data with biological knowledge. Math Biosci 266:65–72
Mingzhu M, Chen A, Wang Z et al (2015) Plant microarray for gene expression profiling and their application. J Agr Technol 11:93–105
Montero-Palmero MB, Martín-Barranco A, Escobar C et al (2014) Early transcriptional responses to mercury: a role for ethylene in mercury-induced stress. New Phytol 201:116–130
Moon J-C, Yim WC, Lim SD et al (2014) Differentially expressed genes and ‘in silico’ analysis in response to ozone (O3) stress of soybean leaves. Austral J Crop Sci 8:276–283
Motamayor JC, Mockaitis K, Schmutz J et al (2013) The genome sequence of the most widely cultivated cacao type and its use to identify candidate genes regulating pod color. Genom Biol 14:r53
Neelam SK, Kim Y-K, Grover A (2014) Coexpression network analysis associated with call of rice seedlings for encountering heat stress. Plant Mol Biol 84:125–143
Park JJ, Yi J, Yoon J et al (2011) OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J 65:194–205
Park HL, Lee SW, Jung KH et al (2013) Transcriptomic analysis of UV-treated rice leaves reveals UV-induced phytoalexin biosynthetic pathways and their regulatory networks in rice. Phytochemistry 96:57–71
Pena LB, Azpilicueta CE, Gallego SM (2011) Sunflower cotyledons cope with copper stress by inducing catalase subunits less sensitive to oxidation. J Trace Elem Med Biol 25:125–129
Qin DD, Xie SC, Liu G et al (2013) Isolation and functional characterization of heat stress responsive gene TaWTF1 with unknown function from wheat. Zhi wu xue Tong Bao 48:31–37
Rashid B, Husnain T, Riazuddin S (2012) Plant genetic engineering: problems and applications. In: Ashraf M, Öztürk M, Ahmad MSA, Aksoy A (eds), Crop production for agricultural improvement. Springer, Dordrecht
Rashid B, Husnain T, Riazuddin S (2014) Genomic approaches and abiotic stress tolerance in plants. In: Ahmad P (ed), Emerging technologies and management of crop stress tolerance, Vol 1. Elsevier, pp 1–26
Rerksiri W, Zhang X, Xiong H et al (2013) Expression and promoter analysis of six heat stress-inducible genes in rice. Sci World J 2013
Sarkar A, Rakwal R, Bhushan AS et al (2010) Investigating the impact of elevated levels of ozone on tropical wheat using integrated phenotypical, physiological, biochemical, and proteomics approaches. J Proteom Res 9:4565–4584
Shaik R, Ramakrishna W (2014) Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol 164:481–495
Shin D, Moon SJ, Han S et al (2011) Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol 155:421–432
Shulaev V, Sargent DJ, Crowhurst RN et al (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43:109–116
Singh A, Pandey A, Baranwal V et al (2012) Comprehensive expression analysis of rice phospholipase D gene family during abiotic stresses and development. Plant Signal Behav 7:847–855
Smits TH, Rezzonico F, Kamber T et al (2010) Complete genome sequence of the fire blight pathogen Erwinia amylovora CFBP 1430 and comparison to other Erwinia spp. Mol Plant-Microb Interac 23:384–393
Takehisa H, Sato Y, Antonio BA et al (2013) Global transcriptome profile of rice root in response to essential macronutrient deficiency. Plant Signal Behav 8:e24409
Theocharis A, Clement C, Barka AE (2012) Physiological and molecular changes in plants grown at low temperatures. Planta 235:1091–1105
Todaka D, Nakashima K, Shinozaki K et al (2012) Toward understanding transcriptional regulatory networks in abiotic stress responses and tolerance in rice. Rice 5:1–9
Ueda Y, Uehara N, Sasaki H et al (2013) Impacts of acute ozone stress on superoxide dismutase (SOD) expression and reactive oxygen species (ROS) formation in rice leaves. Plant Physiol Biochem 70:396–402
Wang X, Tang C, Zhang H et al (2011) TaDAD2, a negative regulator of programmed cell death, is important for the interaction between wheat and the stripe rust fungus. Mol Plant Microb Interac 24:79–90
Wang X, Tang C, Huang X et al (2012) Wheat BAX inhibitor-1 contributes to wheat resistance to Puccinia striiformis. J Exp Bot 63:4571–4584
Wu M, Huang J, Xu S et al (2011) Haem oxygenase delays programmed cell death in wheat aleurone layers by modulation of hydrogen peroxide metabolism. J Exp Bot 62:235–248
Xiansheng W, Liu Y, Jia Y et al (2012) Transcriptional responses to drought stress in root and leaf of chickpea seedling. Mol Biol Rep 39:8147–8158
Xianwen Z, Li J, Liu A et al (2012) Expression profile in rice panicle: insights into heat response mechanism at reproductive stage. PloS One 7:e49652
Xie Y, Cui W, Yuan X et al (2011) Heme Oxygenase-1 is associated with wheat salinity acclimation by modulating reactive oxygen species homeostasis. J Integr Plant Biol 53:653–670
Xie G, Kato H, Imai R (2012) Biochemical identification of the OsMKK6OsMPK3 signalling pathway for chilling stress tolerance in rice1. Biochem J 443:95
Xu Q, Chen L-L, Ruan X et al (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45:59–66
Xue G-P, Way HM, Richardson T et al (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Young ND, Debellé F, Oldroyd GE et al (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524
Zhang C-J, Zhao B-C, Ge W-N et al (2011) An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol 157:1884–1899
Zhang H, Wei L, Miao H et al (2012a) Development and validation of genic-SSR markers in sesame by RNA-seq. BMC Genom 13:316
Zhang Q, Chen W, Sun L et al (2012b) The genome of Prunus mume. Nat Commun 3:1318
Zhang D, Jiang S, Pan J et al (2014) The overexpression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol 16:558–570
Zhang T, Tang J, Sun J (2015) Hex1 related transcriptome of Trichoderma atroviride reveals expression patterns of ABC transporters associated with tolerance to dichlorvos. Biotechnol Lett 37:1421–1429
Zhou ZS, Yang SN, Li H et al (2013) Molecular dissection of mercury responsive transcriptome and sense/antisense genes in Medicago truncatula. J Hazar Mater 252:123–131
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gul, A., Ahad, A., Akhtar, S. et al. Microarray: gateway to unravel the mystery of abiotic stresses in plants. Biotechnol Lett 38, 527–543 (2016). https://doi.org/10.1007/s10529-015-2010-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-2010-2