Abstract
Background
Potassium (K+) is an important nutrient ion in plant cells and plays crucial roles in many plant physiological and developmental processes. In the natural environment, K+ deficiency is a common abiotic stress that inhibits plant growth and reduces crop productivity. Several microarray studies have been conducted on genome-wide gene expression profiles of rice during its responses to various stresses. However, little is known about the transcriptional changes in rice genes under low-K+ conditions.
Results
We analyzed the transcriptomic profiles of rice roots in response to low-K+ stress. The roots of rice seedlings with or without low-K+ treatment were harvested after 6 h, and 3 and 5 d, and used for microarray analysis. The microarray data showed that many genes (2,896) were up-regulated or down-regulated more than 1.2-fold during low-K+ treatment. GO analysis indicated that the genes showing transcriptional changes were mainly in the following categories: metabolic process, membrane, cation binding, kinase activity, transport, and so on. We conducted a comparative analysis of transcriptomic changes between Arabidopsis and rice under low-K+ stress. Generally, the genes showing changes in transcription in rice and Arabidopsis in response to low-K+ stress displayed similar GO distribution patterns. However, there were more genes related to stress responses and development in Arabidopsis than in rice. Many auxin-related genes responded to K+ deficiency in rice, whereas jasmonic acid-related enzymes may play more important roles in K+ nutrient signaling in Arabidopsis.
Conclusions
According to the microarray data, fewer rice genes showed transcriptional changes in response to K+ deficiency than to phosphorus (P) or nitrogen (N) deficiency. Thus, transcriptional regulation is probably more important in responses to low-P and -N stress than to low-K+ stress. However, many genes in some categories (protein kinase and ion transporter families) were markedly up-regulated, suggesting that they play important roles during K+ deficiency. Comparative analysis of transcriptomic changes between Arabidopsis and rice showed that monocots and dicots share many similar mechanisms in response to K+ deficiency, despite some differences. Further research is required to clarify the differences in transcriptional regulation between monocots and dicots.
Similar content being viewed by others
Background
Potassium (K+) is the most abundant monovalent cation in plant cells and accounts for 2–10% of plant dry weight [1]. It plays crucial roles in many physiological processes in living plant cells, including osmoregulation, control of turgor pressure, electrical neutralization, and enzyme activation [1, 2]. The K+ concentration in soil solution is generally within the range of 1 to 200 ppm (approx. 0.025 ~ 5 mM) [3]. In the rhizosphere, the K+ concentration is usually less than 0.3 mM [4]. Therefore, most plants are subjected to low K+ stress at some point during their lives. Most plants can survive under low-K+ conditions, largely because of the high-affinity K+ uptake system in the roots, especially the high-affinity K+ transporters [5]. Different plant species and genotypes have various high-affinity K+ transporters with diverse K+-uptake efficiencies [6].
Previous studies reported that many high-affinity K+ transporter genes are induced by low-K+ conditions. For example, several members of the KUP/HAK/KT family including AtKUP3[7], AtHAK5[8, 9], HvHAK1[10, 11], OsHAK1[12] as well the K+ transporter genes TaAKT1[13] and AtCHX17[14] were induced by K+ starvation. These K+ transporters may be involved in K+ acquisition and homeostasis in plant cells under low-K+ conditions. Furthermore, transcriptomic analysis of Arabidopsis under low-K+ stress identified many candidate genes related to low-K+ perception, and regulatory pathways associated with responses to K+ deficiency [9, 15].
Microarray technology is a convenient tool for rapid analysis of plant gene expression patterns under a variety of environmental conditions. Research on signal transduction pathways activated during the plant response to nutrient deficiency is often initiated from microarray data analyses [16]. For instance, the genome-wide gene expression profiles of rice responses to drought [17], low nitrogen [18], low phosphorus [19–21] and low iron [22] stresses have been analyzed using microarray techniques. However, very little progress has been made in understanding changes in transcription of rice genes under low K+ conditions.
In the present study, we used Affymetrix microarrays to monitor the transcriptomic profiles of rice roots during responses to short- and long-term K+ deficiency. We analyzed the functional categorization of differentially expressed genes and compared the low-K+ response profiles between rice and Arabidopsis. These results will further our understanding of the molecular mechanisms underlying responses to nutrient deficiency in plants.
Results and discussion
K+deficiency inhibits growth of rice seedlings
To investigate the transcriptional changes in rice roots under K+ deficiency, we first determined the appropriate period of low-K+ treatment. Hydroponically grown rice seedlings (2-weeks old) were transferred to K+-free solution (−K) and K+-replete solution (+K, 1 mM K+) as treatment (LK) and control (CK) conditions, respectively. The nutrient solution used was that described by the International Rice Research Institute [23]. After treatment for indicated times (6 h, and 1, 3, 5, and 7 d), the K+-deficient phenotypes of rice seedlings were evaluated. The dry weight and K+ content were also measured at each time point.
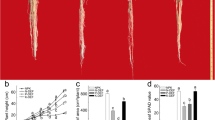
There was no phenotypic difference between K+-starved seedlings and control seedlings after K+ deficiency treatment for 6 h, or 1 or 3 d (Figure 1A). The seedlings showed obvious phenotypic differences after 5 days of K+ starvation, at which time the K+-deficient plants had smaller roots and shoots than those of the control (Figure 1A). The observed phenotypes were consistent with the biomass measurements. Compared with control seedlings, the biomass of roots and shoots of K+-starved seedlings was lower after 5 d K+ deficiency (Figure 1B, 1C). The seedling K+ content showed a decrease after only 6 h, and continued to decrease markedly throughout the K+ deficiency treatment (Figure 1D, E).
K+deficiency inhibits rice growth and affects seedling K+content. (A) Phenotypes of rice seedlings under normal and K+-deficient conditions (CK and LK) at indicated times. (B, C) Dry weight of shoots (B) and roots (C) under CK and LK conditions at indicated times. (D, E) K+ content of shoots (D) and roots (E) at indicated times. Data are means ± SEM (n = 3). Differences between mean values of treatments and controls were compared using t - tests (* P < 0.05, ** P < 0.01).
According to these results, we chose three time points (6 h, and 3 and 5 d) to investigate changes in gene transcription. The seedling K+ content had already decreased at the 6-h time point. It decreased further thereafter, but biomass was not affected until the 3-d time point. At the 5-d time point, the K+ content and biomass were both significantly decreased. Therefore, these three time points may represent different stages of rice root responses to K+ deficiency.
Microarray analysis reveals rice genes showing transcriptional changes in the response to K+deficiency
We used an Affymetrix rice genome array to reveal transcriptional changes during the rice response to K+ deficiency. This array contains 51,279 probes representing two rice cultivars (48,564 japonica transcripts and 1,260 indica transcripts, http://www.affymetrix.com). The root is the main tissue that first senses nutrient deficiency in the environment. Therefore, roots from K+-starved seedlings and controls were harvested at the three time points (6 h, 3 d, and 5 d). Their total RNA was extracted and used for microarray experiments (see “Materials and Methods” for details). To ensure reproducibility and reliability of microarray data, three biological replicates were analyzed at each time point, making a total of 18 rice genome arrays. To assess the reproducibility of microarray data, the correlation coefficients of biological replicates were calculated using GeneSpring GX 11 software. All correlation coefficients were greater than 0.9915 (Additional file 1). We conducted real-time PCR analysis for 12 randomly selected genes to confirm the validity of the microarray data. For most of the 12 genes, their expression patterns in the real-time PCR analysis were similar to those predicted by the rice microarray data (Figure 2). The raw data sets (CEL) and the normalized expression data sets have been deposited in the Gene Expression Omnibus (GSE37161) at the National Center for Biotechnology Information [24] (http://www.ncbi.nlm.nih.gov/geo/).
Confirmation of transcriptomic profiles of selected genes by real-time PCR. LK/CK ratios in microarray data of 12 tested genes compared with real-time PCR results. Sequences of gene-specific primers used for PCR are listed in Additional file 12.
The microarray results showed that approximately 25,000 genes showed positive signals in this data set. This represented approximately 44% of the probe sets on the rice genome array (Additional file 2). Among these expressed genes, we were more interested in those that showed transcriptional changes in response to low-K+ conditions. We analyzed these differentially expressed genes using DNA-Chip Analyzer (dChip) 2007 software [25] (http://www.dchip.org). This revealed genes whose expression levels in K+-starved seedlings were up-regulated or down-regulated more than 1.2-, 1.5-, 2.0-fold (P-value < 0.05) compared with control seedlings (see “Materials and Methods” for details). In total, 2,896, 1,166, and 356 genes showed changes in expression of more than 1.2-, 1.5-, and 2.0-fold, respectively (Additional file 3). Since there were fewer up- or down-regulated genes at the 1.5 or 2.0-fold cutoff thresholds, we used the 1.2-fold cutoff threshold for further analysis in order to contain more candidate genes involving in rice response to K+ deficiency.
The number genes showing changes in expression increased as the K+-starvation time increased from 6 h to 3 d to 5 d (Table 1). At the beginning of the K+-starvation period (6 h and 3 d), there were more up-regulated genes (100 at 6 h, 719 at 3 d) than down-regulated genes (64 at 6 h, 486 at 3 d) (Table 1). This result suggested that many K+-related genes were induced, and that these may be involved in the response and adaption to K+-deficiency during this period. However, at a later stage of K+-starvation (5 d), there were more down-regulated genes (1186) than up-regulated genes (1141) (Table 1). This indicated that the expression of many genes may have been restrained to slow the growth rate of rice, while others were activated to adapt to long-term K+-deficiency.
Previous transcriptomic studies in Arabidopsis showed that only a few genes were transcriptionally regulated by K+ deficiency [9, 15]. In contrast, phosphorus (P) and nitrogen (N) deprivation caused transcriptional changes in a vast number of genes in Arabidopsis[26, 27]. We observed similar trends in rice seedlings. There were fewer genes showing transcriptional changes in the present study than in previous studies on rice seedlings subjected to P- and N-starvation [18–21]. These findings, together with previous data, indicate that post-translational regulation may be more important than transcriptional regulation in plant responses to low-K+ stress [9, 16].
To investigate the similarities and differences among genes showing changes in transcription at different time points during the low-K+ treatment, we carried out a hierarchical cluster analysis and constructed a Venn diagram (Figure 3). The genes showing changes in expression at 3 and 5 d showed similar expression patterns, which were different from those of genes showing changes in expression at 6 h (Figure 3A). This suggested that the genes showing transcriptional changes at 6 h may represent those involved in short-term responses to K+ deficiency, whereas those involved in long-term responses may begin to be activated 3 days later. In addition, some genes showed transcriptional changes at all time points measured—these may have functions throughout the entire period of K+-starvation (Figure 3B, C).
Hierarchical cluster analysis and Venn diagrams of rice genes showing transcriptional changes in response to K+deficiency. (A) Hierarchical cluster analysis of genes showing transcriptional changes at indicated times (6 h, 3 d and 5 d) during K+ deficiency. (B, C) Venn diagrams of up- (B) and down- (C) regulated genes at different LK-treated times. The genes up-regulated at least at one time point were selected for the analysis in (B). Meanwhile, the genes down-regulated at least at one time point were selected for the analysis in (C).
Gene ontology analysis of differentially expressed genes in the rice response to K+deficiency
To evaluate the potential functions of these differentially expressed genes in the response to K+ deficiency, Gene Ontology (GO) analysis was performed using the AgriGO online service [28] (http://bioinfo.cau.edu.cn/agriGO). The 2,896 differentially expressed genes were classified into 13 functional categories including metabolic process (18.8%), membrane (13.5%), cation binding (7.7%), kinase activity (4.2%), transport (3.8%), protein modification (3.7%), and response to stress (3.1%), as shown in Figure 4. Most of these processes were considered to be closely related to the response to K+-deficiency. Further analysis showed that in most of the GO categories, there were far more up-regulated than down-regulated genes at the 6-h and 3-d time points (Additional file 4), but more down-regulated than up-regulated genes at the 5-d time point (Additional file 4). The results of GO analysis may provide some clues to understand the transcriptomic profiles of genes involved in rice responses to K+ deficiency.
Functional category distribution of rice genes showing transcriptional changes in response to K+deficiency. AgriGO web-based tool was used to analyze GO categories of genes showing changes in transcription levels. Percentages of genes showing transcriptional changes in 13 main biological categories are shown.
Metabolic enzymes
Further analysis of differentially expressed genes in the metabolic process category indicated that approximately 37% were related to nitrogen metabolism, 17% to phosphorus metabolism, and 15% to carbohydrate metabolism, together accounting for approximately 70% of genes in this category (Figure 5A). Many metabolic enzymes require K+ ions as a cofactor [29]; therefore, a decrease in cytosolic K+ concentration may influence metabolic processes. A previous study reported that transcriptional regulation of metabolic enzymes may be very important in adaptation to K+ deficiency in Arabidopsis[30].
In this study, the expression levels of two nitrate reductase (NR) genes (Os08g0468100 and Os08g0468700) decreased at a late stage (5 d) of K+ deficiency. Some protein kinases and phospholipases that play roles in phosphorus-related metabolic processes were also transcriptionally regulated. Two PEPCK (phosphoenolpyruvate carboxykinase) genes (Os10g0204400 and Os01g0208700) were significantly down-regulated at the 5 d time point.
Many metabolites in Arabidopsis roots, such as pyruvate, malic acid, and nitrate, significantly decreased when plants were subjected to low-K+ stress [30]. However, 24 h after resupply of K+ in the growth medium, the concentrations of most of these metabolites had recovered [30]. Changes in the cellular concentrations of metabolites may result from K+-induced transcriptional changes in genes encoding metabolic enzymes. Consequently, changes in the K+ supply level would affect their transcription levels. Taken together, these results suggested that long-term K+ deficiency may affect the regulation of metabolic processes, leading to depressed cellular activities and a slower plant growth rate to enable the plant to survive in nutrient-limited conditions.
Cation binding proteins
The differentially expressed genes in the cation binding category mainly encoded proteins that bind iron, zinc, and calcium ions (Figure 5B). We found that approximately one-third of the iron-binding protein genes encoded peroxidases (POXs), which are heme-containing proteins in plant cells [31]. POXs play crucial roles in scavenging reactive oxygen species (ROS) during the processes of cellular metabolism and abiotic stresses [32, 33]. Previous research indicated that accumulation of ROS in plant roots is an early response to K+ deficiency [34, 35], and that many peroxidase genes related to ROS signaling are induced under low-K+ conditions [15, 34]. In the present data, 21 peroxidase genes showed changes in transcription under low-K+ conditions in rice (8 up-regulated and 13 down-regulated). This confirmed the relationship between peroxidases and low-K+ signal sensing in rice. These transcriptional regulated peroxidase genes are classified into six subgroups [36], and listed in Additional file 5.
We also identified nine calcium sensor protein genes showing changes in expression levels during K+ starvation; five were up-regulated (OsCBL5, OsCML1, OsCML18, OsCML20, OsCML31) and four were down-regulated (OsCML5, OsCML11, OsCPK9, OsCCaMK1) (Additional file 5). Previously, Ca2+-binding proteins were reported as being important K+-responsive genes in Arabidopsis[15]. Here, we also identified Ca2+ sensor proteins in rice, which indicated these proteins may connect Ca2+ signaling and down-stream target proteins in the plant response to K+ deficiency.
Kinases and protein modification
In the protein modification category, most genes were identified as kinases (68%) and phosphatases (9%) (Figure 5C). This result suggested that phosphorylation and dephosphorylation may be very important regulatory mechanisms in the rice responses to K+ deficiency. Previous investigations showed that many protein kinases modulated ion transport processes in living cells by regulating the activity of the corresponding ion transporters under ionic stress. This regulatory mechanism is found in both plant and mammalian cells [37–39]. The results of GO analysis in the present study showed that many genes in the kinase and protein modification categories were transcriptionally regulated by K+ starvation (Figure 4). The kinases encoded by these genes may be involved in regulating K+ uptake and K+ homeostasis in rice under K+ deficiency.
According to the Rice Kinase Database [40] (http://phylomics.ucdavis.edu/kinase/index.shtml), there are 1,467 kinase genes corresponding to 1,934 transcripts in the rice genome. These protein kinases can be divided into six major phylogenetic groups; AGC, CAMK, GMGC, CK1, STE, and TKL [40]. In the present study, 123 kinase genes from five groups (all groups except CK1) were transcriptionally regulated by K+ deficiency. This represents 8.4% of the total kinase genes in rice. Among them, most changed kinase genes belong to TKL group (Figure 6A). TKL group mainly consists of receptor-like protein kinases. Many of them have been reported to play crucial roles in plant response to various external signals [41]. For example, the leucine-rich repeat (LRR) receptor-like protein kinases BRI1 and BAK1 are involved in brassinosteroid signal transduction in Arabidopsis[42, 43]. In addition, RPK1 participates in ABA signaling pathway [44]. Therefore, we hypothesized that the transcriptionally changed genes in TKL group may be also involved in rice response to low-K+ stress. These proteins possibly sense external low-K+ signal and transduce it to downstream components via phosphorylation.
We also analyzed the percentages of kinase genes showing changes in transcription in each kinase group. A greater proportion of the kinases in the CAMK and STE groups (15.6% and 12%, respectively) showed changes in transcription compared with the other three groups (Figure 6B). This result indicated that kinases in these two groups may be more important in rice responses to K+ deficiency. In the CAMK group, the genes showing changes in transcription included ten OsCIPKs, two OsSnRK1-type, two OsCPKs, and one OsCCaMK (Table 2). In the STE group, four OsWNK protein genes and two MAP3K kinase genes showed transcriptional changes (Table 2). In our microarray data, many kinase genes from OsCIPK (CBL-interacting protein kinase) and OsWNK (with no K = lysine) families were transcriptionally regulated in rice seedlings subjected to K+ deficiency (Additional file 6, 7).
CIPKs are plant-specific serine/threonine protein kinases that form protein complexes with calcium sensor CBLs (calcineurin B-like proteins) to perform functions [45, 46]. A previous investigation in Arabidopsis showed that the protein kinase CIPK23 together with CBL1/9 activates AKT1-mediated K+ uptake in roots under low-K+ conditions [47, 48]. Another recent study found that CIPK6 modulates the translocation of the K+ channel, AKT2, to the plasma membrane by interacting with CBL4, and enhances AKT2 activity at the plasma membrane [49]. Rice OsCIPKs show high similarity to Arabidopsis CIPKs in their amino acid sequence; therefore, similar regulatory mechanisms may also exist in rice. According to the sequences of AtCIPKs in Arabidopsis, Kolukisaoglu et al. (2004) identified at least 30 OsCIPKs from rice genome [50]. In NCBI and RGAP database, we can find 33 OsCIPKs in the updated annotations. In Affymetrix rice microarray, these 33 OsCIPKs genes are represented by 31 probe sets. Approximately one-third of OsCIPKs showed high expression levels in rice roots (average signal intensity values ≥ 500). In the present data, eight OsCIPK genes (OsCIPK2, 6, 9, 10, 14/15, 23, 26) were up-regulated and two OsCIPK genes (OsCIPK29, 31) were down-regulated under low-K+ stress (Table 2, Additional file 6). Increased expressions of these kinase genes may regulate activity of the K+ channel, enhancing K+ uptake under K+-deficient conditions.
WNK is another kind of serine/threonine protein kinase, which was first investigated in mammals [51]. The WNKs in mammals function in renal regulation of ion transport, especially in Na+ and K+ transport [39, 52]. In higher plants, WNK homologs were identified in several plant species such as Arabidopsis (10 genes) and rice (9 genes) [53–55]. A recent report showed that AtWNK8 could phosphorylate the subunit of vacuolar H+-ATPase and regulate ion transport in Arabidopsis[56]. In rice, OsWNK1 is regulated by abiotic stress and plays a role in the internal circadian rhythm [55]. In our microarray experiments, five OsWNKs were strongly expressed in rice roots (average signal intensity values ≥ 500). In addition, OsWNK5, 7, 8 were up-regulated and OsWNK9 was down-regulated under low-K+ conditions (Table 2, Additional file 7).
Some kinases from these two families phosphorylate ion transporters and regulate ion transport in plant cells. It is likely that the expression levels of these kinase genes increased in rice seedlings under low-K+ conditions to regulate ion transporters, thereby enhancing transporter-mediated K+-uptake in rice roots.
Membrane proteins and ion transporters
Uptake of K+ in plant cells is primarily mediated by K+ transporters and K+ channels located at the plasma membrane [57–59]. K+ transporters are major components of the high-affinity K+-uptake machinery that operates under low external K+ conditions [5, 60, 61]. Many high-affinity K+ transporter genes from different plant species are induced by low-K+ stress, such as AtHAK5, HvHAK1, and OsHAK1[8–12]. The increase in expression levels of genes encoding K+ transporters may be a rapid and direct strategy for plants to increase K+-uptake and overcome K+ deficiency. This appears to be a common regulatory mechanism among different plant species.
In the rice genome, the major K+ transporters are derived from OsHAK (27 genes) and OsHKT (9 genes) families [62, 63]. In our microarray data, three OsHAK genes (1, 7 and 11) were markedly up-regulated under low-K+ stress (Table 2, Additional file 8). This is consistent with a previous report that K+ starvation induced expression of OsHAK1[12]. We speculated that these K+ transporters may be involved in K+ uptake under low-K+ conditions.
For the OsHKT genes, all members of the OsHKT family were expressed at low levels in rice roots (Additional file 8). Only OsHKT2;1 was strongly up-regulated during K+ deficiency (Table 2). This result is consistent with reports that homologs of OsHKT2;1 in wheat and barley were significantly induced under low-K+ conditions [64, 65]. It is interesting that this gene product was reported as a Na+ transporter, not a K+ transporter, when identified in heterologous expression systems [65, 66]. In K+-starved rice roots, OsHKT2;1 increased the concentration of Na+ ions, so that these Na+ ions could serve as nutritional ions and enhance growth of rice seedlings under low-K+ conditions [67].
Besides K+ transporter genes, we found that the transcriptional levels of some phosphate and nitrate transporter genes changed in rice roots under K+-deficiency. In our microarray data, two phosphate transporter genes (OsPHO1;1 and OsPHT1;4) were markedly up-regulated after 5 d of K+ starvation (Table 2). In contrast, two nitrate transporter genes (Os02g0112600 and Os02g0689900) were down-regulated at the same time (Table 2). A similar regulatory mechanism was also found in Arabidopsis. The expression levels of three Arabidopsis nitrate transporter genes (AtNRT2;1, AtNRT2;3, AtNRT2;6) were reduced under low-K+ conditions and quickly increased after K+ resupply [15]. In contrast, P- and N-deficiencies increased the expressions of K+ transporters such as HAK5, KUP10, KUP11 in Arabidopsis[35, 68]. These findings indicate that K, P, and N nutrient uptakes are inter-related. The deficiency of one type of nutrient ion may affect the absorption and translocation of other two types of ions in plants. This latent mechanism could facilitate the homeostasis of nutrient ions in plant cells. However, further research is required to clarify the details of this regulatory mechanism.
Comparative analysis of low-K+ responses between rice and Arabidopsis
Rice and Arabidopsis are both model plants, the former being a model monocot and the latter a model dicot. It is meaningful to compare transcriptomic changes between these two species under low-K+ stress to understand which mechanisms are shared and which are specific to each plant, and to gain insight into common regulatory mechanisms. Previous investigations have indicated that many genes are transcriptionally regulated under P- and N-deficient conditions; this was observed in both Arabidopsis[26, 27, 68] and rice [18–21]. In contrast, fewer genes were induced by K+ deficiency in both Arabidopsis[9, 15] and rice (present data; Additional file 9). These findings suggest that transcriptional regulation is probably more important in plant responses to low P and N stress than in the plant response to low-K+ stress.
To analyze transcriptomic differences between rice and Arabidopsis under low-K+ stress, we compared microarray data from Arabidopsis[9] with that in the present study. We reanalyzed their microarray data (four Arabidopsis ATH1 microarrays, two time points, no biological duplication) with the same data processing methods (1.2-fold cutoff threshold) used in this study. Then, the results of GO analysis from rice and Arabidopsis were compared (Additional file 10). Generally, the genes showing transcriptional changes in rice and Arabidopsis displayed similar GO distribution patterns in their responses to low-K+ stress. This result indicated that genes in the categories of metabolic process, membrane, and cation binding may play crucial roles in responses to low-K+ stress in both rice and Arabidopsis. However, there were also some differences. There was a greater proportion of genes related to the stress response and to development in Arabidopsis than in rice (Additional file 10), suggesting that these genes were more important in the response to low-K+ stress in Arabidopsis.
In present data, we found 33 hormone-related genes that respond to K+ deficiency in rice roots. Most of them (23) are related to auxin, including 10 OsIAAs, 4 OsSUARs and 2 OsGHs etc. These genes have been listed in “Additional file 11”. The transcriptional levels of several members from these gene families can be induced by auxin [69]. While, they are also up-regulated or down-regulated under low K+ conditions in rice roots, which indicates the link between auxin and K+ nutrient signaling. It has been reported that auxin is accumulated in central cylinder cells in the distal elongation zone of Arabidopsis roots after K+ deprivation [70]. Therefore it is hypothesized that auxin may also generate in rice roots after K+ deprivation and regulate rice root morphology under K+-deficient conditions. Armengaud et al., (2004) reported that many JA-related genes were induced after K+ deprivation, and highlighted a novel role of JA in Arabidopsis K+ nutrient signaling. However, it was noteworthy that these JA-related genes were mainly induced in Arabidopsis shoots, but less in roots. Therefore, it is suggested that different phytohormones may play roles in different organs to facilitate plant adapting K+-deficient stress.
These transcriptomic comparisons may provide some clues to understand differences between rice and Arabidopsis in their responses to low-K+ stress. However, further research is required to explore the differences in transcriptional regulation under nutrient deficiency between monocots and dicots.
Conclusions
K+ deficiency is a common abiotic stress in the natural environment, and it inhibits plant growth and reduces crop production. In the present study, we analyzed Affymetrix microarrays to study the transcriptomic profiles of rice roots during the response to K+ deficiency. The microarray data showed that fewer genes are involved in the response to K+ deficiency than in the responses to P and N-deficiencies. Therefore, transcriptional regulation is probably more important in the plant response to low P- and low N-stress than to low-K+ stress. In total, 2,896 genes showed significant changes in transcription levels during low-K+ treatment. These were mainly in the GO categories of metabolic process, membrane, cation binding, kinase activity, transport, and so on. Many genes in protein kinase and ion transporter families were markedly up-regulated, suggesting that they may play important roles in rice responses to K+ deficiency. We also conducted a comparative analysis of transcriptomic changes between Arabidopsis and rice under low-K+ stress. The result indicated that monocots and dicots may share similar mechanisms in their responses to K+ deficiency, despite some differences. However, further research is required to clarify the differences in transcriptional regulation between monocots and dicots.
Methods
Plant growth condition and low-K+stress treatment
Rice cultivar Nipponbare (Oryza sativa L. ssp. japonica) plants were cultured hydroponically in a growth chamber under the following conditions: relative humidity, 50–70%; 12-h light/12-h dark photoperiod; temperatures, 30°C days, 25°C nights.
The composition of the nutrient solution was as described by the International Rice Research Institute [23]. The solution contained 1.427 mM NH4NO3, 0.323 mM NaH2PO4·2H2O, 0.512 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4·7H2O, 9.474 μM MnCl2·4H2O, 0.075 μM (NH4)6Mo7O24·4H2O, 18.882 μM H3BO3, 0.152 μM ZnSO4·7H2O, 0.155 μM CuSO4·5H2O, 0.031 mM FeSO4·7H2O, and 0.031 mM Na2EDTA·2H2O, (pH = 5.5). The nutrient solution was replaced every second day. First, the rice seedlings were cultured in normal solution for two weeks. Then, half of them were transferred to -K nutrient solution (lacking K2SO4) as the K+-deficient treatment (LK), and another half were transferred to normal solution (+K) as control (CK). Roots and shoots of both control and treated seedlings were collected at indicated times for further assays.
Biomass and K+content measurement
The roots and shoots of control and treated rice seedlings were collected at various times during the K+-deficiency treatment (6 h, and 1, 3, 5, and 7 d). The root and shoot tissues were harvested separately, dried at 80°C for 48 h, and then weighed (dry weight). To measure K+ content, dry samples were incinerated in a muffle furnace at 575°C for 5 h, the residue was dissolved in 0.1 N HCl, and the K+ content was measured by atomic absorption spectrophotometry (Hitachi Z-5000). Three biological replicates were used for phenotypic tests, biomass and K+ content measurements. The t - test (* P < 0.05, ** P < 0.01) was used to analyze the statistical significance.
RNA isolation, target preparation, and hybridization to Affymetrix genechips
Fresh roots of both control and treated rice seedlings were collected at indicated times during the K+-deficiency treatment (6 h, 3 and 5 d). The roots were immediately frozen in liquid nitrogen and stored at −80°C until further RNA extraction. Every RNA sample was derived from five independent seedlings. Total RNA was extracted from rice roots using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Quality characterization of RNA samples was determined and confirmed using a NanoDrop 2000 fluorospectrometer and formaldehyde denaturing gel electrophoresis. Then, the total RNA was purified using the RNeasy Mini kit (Qiagen, Valencia, CA, USA). The purified RNA was used to generate first-strand cDNA in a reverse transcription reaction containing One-Cycle Target Labeling and Control reagents (Affymetrix, Santa Clara, CA, USA) at 42°C for 2 h and 16°C for 1 h. The double-stranded cDNAs were used to generate cRNA via an in vitro transcriptional reaction. The cRNA was labeled with biotin, and 20 μg labeled cRNA was fragmented. The size distribution of cRNAs and fragmented cRNAs was assessed using an Eppendorf Biophotometer and electrophoresis. Fragmented cRNA (15 μg) was added to 300 μL hybridization solution, and then 200 μL of this mixture was used for hybridization on Affymetrix rice genome arrays for 16 h at 45°C. The standard wash and double-stain protocols were applied using a fluidics station (Affymetrix GeneChip Fluidics Station 450). The arrays were scanned on a genechip scanner (Affymetrix GeneChip Scanner 3000).
Microarray data processing and analysis
First, the scanned arrays were analyzed using Affymetrix GCOS 1.0 (MAS 5.0) software to generate raw data, which were saved as CEL files. Then, DNA-Chip Analyzer (dChip) 2007 software [25] (http://www.dchip.org) was used to normalize all 18 array sets. A gene was considered to be expressed when it had detection call “P” in all three replicates, and when its detection P-value was less than 0.05. We used GeneSpring GX 11 software to analyze the correlation coefficients between every pair of arrays. The genes showing changes in transcription in response to K+ deficiency were analyzed using dChip 2007 software. A gene was considered to show transcriptional changes when it satisfied two conditions: (1) up-regulation or down-regulation more than 1.2-fold after K+-deficiency treatment; (2) a fold-change P-value of less than 0.05. A negative or positive value represents downward or upward regulation, respectively. We conducted hierarchical cluster analysis of genes showing transcriptional changes using TIGR MeV 4.2 software [71] (http://www.tm4.org). The genes up- or down-regulated at least at one time point were collected respectively, which were used for Venn diagram analysis. The AgriGO web service (http://bioinfo.cau.edu.cn/agriGO/analysis.php) was used to analyze the functional categories of genes showing transcriptional changes. Microsoft Excel and Access were used to extract and manage microarray data.
Real-time PCR analysis
The RNA samples for microarray experiment (three biological replicates) were also used for real-time PCR assays in order to ensure the reliability and repeatability of the results. To eliminate genomic DNA contamination, total RNA was treated with DNase I (RNase Free) (Takara, Dalian, China). Then, the total RNA was used to synthesize cDNA in a reverse transcription reaction using random primers (Promega, Madison, WI, USA). The cDNA samples were diluted to 4 ng/μL. Three biological replicates were performed using the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on a 7500 Real Time PCR System machine (Applied Biosystems) according to the manufacturer’s protocols. The gene-specific primers (Additional file 12) were designed using Primer 3 (http://frodo.wi.mit.edu/primer3/input.htm) and DNAMAN software. Amplification of 18S rRNA was used as an internal control to normalize the data.
References
Leigh RA, Wyn Jones RG: A hypothesis relating critical potassium concentrations for growth to the distribution and function of this ion in the plant cell. New Phytol. 1984, 97: 1-13. 10.1111/j.1469-8137.1984.tb04103.x.
Clarkson DT, Hanson JB: The mineral nutrition of higher plants. Annu Rev Plant Phsyiol. 1980, 31: 239-298. 10.1146/annurev.pp.31.060180.001323.
Barber SA: In Soil Nutrient Bioavailability: A Mechanistic Approach. Potassium. 1995, Edited by Barber SA, New York: Wiley, 231-232.
Schroeder JI, Ward JM, Gassmann W: Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct. 1994, 23: 441-471. 10.1146/annurev.bb.23.060194.002301.
Epstein E, Rains DW, Elzam OE: Resolution of dual mechanisms of potassium absorption by barley roots. PNAS. 1963, 49: 684-692. 10.1073/pnas.49.5.684.
Pettersson S, Jensen P: Variation among species and varieties in uptake and utilization of potassium. Plant Soil. 1983, 72: 231-237. 10.1007/BF02181962.
Kim EJ, Kwak JM, Uozumi N, Schroeder JI: AtKUP1: An Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell. 1998, 10: 51-62.
Ahn SJ, Shin R, Schachtman DP: Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 2004, 134: 1135-1145. 10.1104/pp.103.034660.
Gierth M, Maser P, Schroeder JI: The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 2005, 137: 1105-1114. 10.1104/pp.104.057216.
Santa-María GE, Rubio F, Dubcovsky J, Rodriguez-Navarro A: The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell. 1997, 9: 2281-2289.
Fulgenzi FR, Peralta ML, Mangano S, Danna CH, Vallejo AJ, Puigdomenech P, Santa-María GE: The ionic environment controls the contribution of the barley HvHAK1 transporter to potassium acquisition. Plant Physiol. 2008, 147: 252-262. 10.1104/pp.107.114546.
Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A: Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 2002, 130: 784-795. 10.1104/pp.007781.
Buschmann PH, Vaidyanathan R, Gassmann W, Schroeder JI: Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 2000, 122: 1387-1397. 10.1104/pp.122.4.1387.
Cellier F, Conejero G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F: Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J. 2004, 39: 834-846. 10.1111/j.1365-313X.2004.02177.x.
Armengaud P, Breitling R, Amtmann A: The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 2004, 136: 2556-2576. 10.1104/pp.104.046482.
Schachtman DP, Shin R: Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol. 2007, 58: 47-69. 10.1146/annurev.arplant.58.032806.103750.
Wang D, Pan YJ, Zhao XQ, Zhu LH, Fu BY, Li ZK: Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics. 2011, 12: 149-10.1186/1471-2164-12-149.
Lian X, Wang S, Zhang J, Feng Q, Zhang L, Fan D, Li X, Yuan D, Han B, Zhang Q: Expression profiles of 10,422 genes at early stage of low nitrogen stress in rice assayed using a cDNA microarray. Plant Mol Biol. 2006, 60: 617-631. 10.1007/s11103-005-5441-7.
Wasaki J, Yonetani R, Kuroda S, Shinano T, Yazaki J, Fujii F, Shimbo K, Yamamoto K, Sakata K, Sasaki T, Kishimoto N, Kikuchi S, Yamagishi M, Osaki M: Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ. 2003, 26: 1515-1523. 10.1046/j.1365-3040.2003.01074.x.
Wasaki J, Shinano T, Onishi K, Yonetani R, Yazaki J, Fujii F, Shimbo K, Ishikawa M, Shimatani Z, Nagata Y, Hashimoto A, Ohta T, Sato Y, Miyamoto C, Honda S, Kojima K, Sasaki T, Kishimoto N, Kikuchi S, Osaki M: Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J Exp Bot. 2006, 57: 2049-2059. 10.1093/jxb/erj158.
Li L, Liu C, Lian X: Gene expression profiles in rice roots under low phosphorus stress. Plant Mol Biol. 2009, 72: 423-432.
Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, Shou H: Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 2009, 151: 262-274. 10.1104/pp.109.141051.
Yoshida S, Forno DA, Cook JH, Gomez KA: Laboratory manual for physiological studies of rice. 1976, Manila: International Rice Research Institute, 61-67.
Edgar R, Domrachev M, Lash AE: Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30: 207-210. 10.1093/nar/30.1.207.
Parmigiani G, Garret E, Irizarry R, Seger S, et al: The analysis of gene expression data: methods and software. 2003, New York: Springer: Edited by Giovanni Parmigiani, 120-139.
Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW: Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003, 132: 1260-1271. 10.1104/pp.103.021022.
Wang R, Okamoto M, Xing X, Crawford NM: Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132: 556-567. 10.1104/pp.103.021253.
Du Z, Zhou X, Ling Y, Zhang Z, Su Z: AgriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38: W64-W70. 10.1093/nar/gkq310.
Wyn Jones RG, Pollard A: In Inorganic Plant Nutrition. Protein, enzymes and inorganic ions. 1983, Berlin: Springer: Edited by Läuchli A and Bieleski RL, 528-562.
Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y: Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 2009, 150: 772-785. 10.1104/pp.108.133629.
Duroux L, Welinder K: The peroxidase gene family in plants: a phylogenetic overview. J Mol Evol. 2003, 57: 397-407. 10.1007/s00239-003-2489-3.
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F: Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9: 490-498. 10.1016/j.tplants.2004.08.009.
Lüthje S, Meisrimler CN, Hopff D, Möller B: Phylogeny, topology, structure and functions of membrane-bound class III peroxidases in vascular plants. Phytochemistry. 2011, 72: 1124-1135. 10.1016/j.phytochem.2010.11.023.
Shin R, Schachtman DP: Hydrogen peroxide mediates plant root cell response to nutrient deprivation. PNAS. 2004, 101: 8827-8832. 10.1073/pnas.0401707101.
Shin R, Berg RH, Schachtman DP: Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 2005, 46: 1350-1357. 10.1093/pcp/pci145.
Passardi F, Longet D, Penel C, Dunand C: The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004, 65: 1879-1893. 10.1016/j.phytochem.2004.06.023.
Luan S: The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14: 37-42. 10.1016/j.tplants.2008.10.005.
Weinl S, Kudla J: The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 2009, 184: 517-528. 10.1111/j.1469-8137.2009.02938.x.
McCormick JA, Ellison DH: The WNKs: a typical protein kinases with pleiotropic actions. Physiol Rev. 2011, 91: 177-219. 10.1152/physrev.00017.2010.
Dardick C, Chen J, Richter T, Ouyang S, Ronald P: The rice kinase database. A phylogenomic database for the rice kinome. Plant Physiol. 2007, 143: 579-586.
Morris ER, Walker JC: Receptor-like protein kinases: the keys to response. Curr Opin Plant Biol. 2003, 6: 339-342. 10.1016/S1369-5266(03)00055-4.
Li J, Chory J: A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997, 90: 929-938. 10.1016/S0092-8674(00)80357-8.
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC: BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002, 110: 213-222. 10.1016/S0092-8674(02)00812-7.
Osakabe Y, Maruyama K, Seki M, Satou M, Shinozaki K, Yamaguchi-Shinozaki K: Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005, 17: 1105-1119. 10.1105/tpc.104.027474.
Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J: Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell. 1999, 11: 2393-2405.
Kim KN, Cheong YH, Gupta R, Luan S: Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 2000, 124: 1844-1853. 10.1104/pp.124.4.1844.
Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH: A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006, 125: 1347-1360. 10.1016/j.cell.2006.06.011.
Li L, Kim BG, Cheong YH, Pandey GK, Luan S: A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. PNAS. 2006, 103: 12625-12630. 10.1073/pnas.0605129103.
Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K: Corratge Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, Kudla J: Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 2011, 21: 1116-1130. 10.1038/cr.2011.50.
Kolukisaoglu U, Weinl S, Blazevic D, Batistič O, Kudla J: Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004, 134: 43-58. 10.1104/pp.103.033068.
Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH: WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000, 275: 16795-16801. 10.1074/jbc.275.22.16795.
Kahle KT, Ring AM, Lifton RP: Molecular physiology of the WNK kinases. Annu Rev Physiol. 2008, 70: 329-355. 10.1146/annurev.physiol.70.113006.100651.
Nakamichi N, Murakami-Kojima M, Sato E, Kishi Y, Yamashino T, Mizuno T: Compilation and characterization of a novel WNK family of protein kinases in Arabidopsis thaliana with reference to circadian rhythms. Biosci Biotech Bioch. 2002, 66: 2429-2436. 10.1271/bbb.66.2429.
Wang Y, Liu K, Liao H, Zhuang C, Ma H, Yan X: The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biol. 2008, 10: 548-562. 10.1111/j.1438-8677.2008.00072.x.
Kumar K, Rao KP, Biswas DK, Sinha AK: Rice WNK1 is regulated by abiotic stress and involved in internal circadian rhythm. Plant Signal Behav. 2011, 3: 316-320.
Hong-Hermesdorfa A, Brüxa A, Grüberb A, Grüberb G, Schumacher K: A WNK kinase binds and phosphorylates V-ATPase subunit C. FEBS Lett. 2006, 580: 932-939. 10.1016/j.febslet.2006.01.018.
Véry AA, Sentenac H: Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol. 2003, 54: 575-603. 10.1146/annurev.arplant.54.031902.134831.
Ashley MK, Grant M, Grabov A: Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot. 2006, 57: 425-436.
Ward JM, Mäser P, Schroeder JI: Plant ion channels: gene families, physiology, and functional genomics analyses. Annu Rev Physiol. 2009, 71: 59-82. 10.1146/annurev.physiol.010908.163204.
Maathuis FJ, Ichida AM, Sanders D, Schroeder JI: Roles of higher plant K+ channels plant. Plant Physiol. 1994, 114: 1141-1149.
Britto DT, Kronzucker HJ: Cellular mechanisms of potassium transport in plants. Physiol Plantarum. 2008, 133: 637-650. 10.1111/j.1399-3054.2008.01067.x.
Amrutha RN, Sekhar PN, Varshney RK, Kishor PBK: Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 2007, 172: 708-721. 10.1016/j.plantsci.2006.11.019.
Gupta M, Qiu X, Wang L, Xie W, Zhang C, Xiong L, Lian X, Zhang Q: KT/HAK/KUP potassium transporters gene family and their whole-life cycle expression profile in rice (Oryza sativa). Mol Genet Genomics. 2008, 280: 437-452. 10.1007/s00438-008-0377-7.
Wang TB, Gassmann W, Rubio F, Schroeder JI, Glass ADM: Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 1998, 118: 651-659. 10.1104/pp.118.2.651.
Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A: Sodium transport and HKT transporters: the rice model. Plant J. 2003, 34: 788-801. 10.1046/j.1365-313X.2003.01764.x.
Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A: Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001, 27: 129-138. 10.1046/j.1365-313x.2001.01077.x.
Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI: Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 2007, 26: 3003-3014. 10.1038/sj.emboj.7601732.
Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC: A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. PNAS. 2005, 102: 11934-11939. 10.1073/pnas.0505266102.
Abel and Theologis: Early Genes and Auxin Action. Plant Physiol. 1996, 111: 9-17. 10.1104/pp.111.1.9.
Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A: Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. Plant J. 2004, 40: 523-535. 10.1111/j.1365-313X.2004.02230.x.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J: TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003, 34: 374-378.
Acknowledgements
We thank Dr. Zhen Su (China Agricultural University) for critical discussions about data analysis. This work was supported by the National Basic Research Program of China (973 Program, Grant No. 2011CB100300), and the National Natural Science Foundation of China (Grant No. 30900269 to WY).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WY conceived the study. MTL carried out phenotypic tests and physiological analyses. MTL carried out RNA extractions, performed real-time RT-PCR analyses, and drafted the manuscript. WY and MTL designed the experiments and analyzed microarray data. WY and WWH supervised the project and finalized the paper. All authors read and approved the final manuscript.
Electronic supplementary material
12870_2012_1102_MOESM4_ESM.pdf
Additional file 4: Functional classification of genes showing transcriptional changes (up-regulated and down-regulated) at 6 h (A), 3 d (B), and 5 d (C) of K+deficiency. AgriGO web-based tool was used to analyze GO categories of differentially expressed genes. Numbers of genes showing transcriptional changes in 13 main biological categories are shown. (PDF 75 KB)
12870_2012_1102_MOESM5_ESM.xls
Additional file 5: Genes encoding calcium sensors and peroxidase proteins showing transcriptional changes during K+-deficiency treatment.(XLS 60 KB)
12870_2012_1102_MOESM9_ESM.pdf
Additional file 9: Comparison of differentially expressed genes between rice and Arabidopsis in responses to K+deficiency.(PDF 91 KB)
12870_2012_1102_MOESM10_ESM.pdf
Additional file 10: Comparison of GO classifications between rice and Arabidopsis in responses to K+deficiency.(PDF 72 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ma, TL., Wu, WH. & Wang, Y. Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol 12, 161 (2012). https://doi.org/10.1186/1471-2229-12-161
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-12-161