Abstract
The SnRK1 complex in plants regulates metabolism in response to environmental stresses and glucose depletion, for stress adaptation and energy homeostasis. Through phosphorylation of various targets, SnRK1 orchestrates intricate regulatory mechanisms involved in autophagy, nutrient remobilization, and TOR activity inhibition, showcasing its pivotal role in coordinating plant metabolism and stress responses. The present study aimed to identify members of the SnRK1 gene family in the maize genome and characterize them using bioinformatics and expression analyses under aphid feeding, drought, and cold stress. The focus of the study was to conduct a comprehensive analysis towards determining gene diversity of ZmSnRK1 genes, constructing intricate 3D structures, and identifying stress-related cis-elements. Four SnRK1 genes were identified, which were named ZmSnRK1.1, ZmSnRK1.2, ZmSnRK1.3, and ZmSnRK1.4. The SnRK1 proteins were found to have a distribution of conserved motifs; however, the distinction between monocots and dicots in the phylogenetic tree was clearly demonstrated. Analysis of the promoter region revealed that the ZmSnRK1 genes contain stress-related cis-elements. Compared to the control, ZmSnRK1.3 significantly upregulated in response to aphid feeding and cold stress, while ZmSnRK1.2 showed elevated expression under drought conditions. The expression of the other two genes under these treatments was generally unperturbed. The findings of this study are poised to establish a valuable scientific foundation for future research on the roles of the SnRK1 gene family in plants, providing valuable insights for enhancing genetic resilience to stress and optimizing yield traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants adjust their metabolisms when exposed to biotic and abiotic environmental stresses (Wurzinger et al. 2018) The process of phosphorylation and dephosphorylation of proteins is crucial for plants to transmit the environmental stress signals into biological effects (Cohen 1988). Research shows that protein kinases, through membrane receptor proteins, are important regulators that perceive environmental signals and activate different protein phosphorylation pathways (Hunter 1995). The AMPK/SNF1/SnRK1 complex, known as an essential component of the evolutionarily conserved energy management network, is one of the important protein kinases that mediates the regulation of cellular energy metabolism (Alderson et al. 1991; Broeckx et al. 2016; Celenza and Carlson 1986). In higher eukaryotes, AMPK/SNF1/SnRK1 protein kinases (PK) function as key cellular energy sensors initiating adaptive changes necessary to maintain the energy homeostasis (Broeckx et al. 2016). SNF1, also known as Sucrose Non-Fermenting 1, was found in Saccharomyces cerevisiae and characterized as the Ser/Thr kinase (Carlson et al. 1981). SnRK1 is widely reported in plants as the ortholog of SNF1, along with its paralogs such as SnRK2 and SnRK3 (Halford and Grahame 1998; Hrabak et al. 2003).
SnRK1 kinases function as heterotrimeric complexes consisting of highly conserved catalytic α subunits and regulatory β and γ subunits (Celenza and Carlson 1986; Polge and Thomas 2007). It is rapidly activated in response to decreasing levels of glucose, a preferred carbon (C) source that can be rapidly fermented (Hedbacker and Carlson 2008). In mammals, the AMPK, once activated by energy shortage, restores energy homeostasis by activating ATP-generating catabolic pathways such as glycolysis and fatty acid oxidation, while inhibiting ATP-consuming biosynthetic and other growth processes (Hardie et al. 2012a, b). Similarly, in plants and yeast, SnRK1 and SNF1 kinases, respectively, emerge as a central energy sensor in the metabolic signaling network controlling important events such as plant growth, development and stress tolerance (Hulsmans et al. 2016; Lastdrager et al. 2014; Pathak et al. 2022; Smeekens et al. 2010; Xiong and Sheen 2015). SnRK1 can be activated under aphid feeding, drought or cold conditions that directly or indirectly cause energy deficit, affecting processes like photosynthesis, respiration or carbon allocation (Broeckx et al. 2016). Additionally, SnRK1 triggers autophagy and nutrient remobilization by phosphorylating ATG1 (Chen et al. 2017), and inhibits TOR activity, another energy sensor that responds to the availability of nutrients, by phosphorylating RAPTOR (Rodriguez et al. 2019). These intricate regulatory mechanisms underscore SnRK1’s multifaceted role in plant metabolism, growth, development, and stress responses. Furthermore, it acts as a central regulator, activating metabolic pathways crucial for maintaining metabolic balance, especially in stress conditions (Halford and Hey 2009).
Given the substantial role of SnRK1 in regulating metabolic and stress signaling pathways, our study focused on identifying SnRK1 members in maize. We have conducted comprehensive analyses including determining gene diversity levels of ZmSnRK1 genes, analyzing their domain structures, constructing intricate 3D structures with cavity pockets, and identifying stress-related cis-elements. It extends beyond the typical scope of research by encompassing all varieties of maize SnRK genes (Feng et al. 2022). Notably, SnRK1 kinases serve as crucial metabolic sensors, orchestrating responses to various stressors and maintaining energy balance through direct phosphorylation of key metabolic enzymes and regulatory proteins, alongside extensive transcriptional reprogramming. Hence, we detected SnRK1 expression profiles under aphid feeding, drought, or cold stress, offering valuable insights for enhancing genetic resilience to stress and optimizing yield traits.
Materials and methods
Genome-wide identification of ZmSnRK1 genes
First, the UniProtKB database (https://www.uniprot.org/) was used to obtain SnRK1 protein sequences from Arabidopsis thaliana (Q38997: SNRK1.1 and P92958: SNRK1.2) and Oryza sativa (Q852Q2: SNRK1A and Q852Q1: SNRK1B). These reference sequences were then used to find orthologs of the SnRK1 genes in the maize genome by means of blastp analyses in the Phytozome v13 database (https://phytozome-next.jgi.doe.gov/). The reliability of the retrieved maize SnRK1 sequences was further checked by domain analysis using the SMART (http://smart.embl-heidelberg.de/) and the InterPro (https://www.ebi.ac.uk/interpro/search/sequence/) online servers.
Sequence analyses
The physico-chemical features of the SnRK1 protein sequences were predicted by ProtParam tool (http://web.expasy.org/protparam/; Gasteiger et al. 2005), and the subcellular localization was predicted by WoLF PSORT (advanced protein subcellular localization prediction tool) (https://wolfpsort.hgc.jp/) (Horton et al. 2007). Exon–intron structures of SnRK1 genes were obtained using the Phytozome database v13 (https://phytozome-next.jgi.doe.gov/), and percent identity (%) was calculated using blastp tool in NCBI (https://www.ncbi.nlm.nih.gov/). ZmSnRK1 gene diversity level and Tajima’s D were calculated using the MEGA11 software (Tamura et al. 2021).
Conserved motif and phylogenetic analyses
The conserved motif structures of the SnRK1 sequences were found using the multiple em for motif elicitation (MEME) tool v5.5.5 (http://meme-suite.org/tools/meme) with the following parameter settings: maximum number of motifs to find as 10; minimum motif width as 6, and maximum motif width as 50 (Bailey et al. 2009). The phylogenetic tree was generated by MEGA11 (Tamura et al. 2021) using Neighbor-Joining (NJ) method for 1000 bootstraps to infer evolutionary history. Evolutionary distances were calculated using the Poisson correction method. All ambiguous positions were removed for each sequence pair (pairwise deletion option).
Co-expression network and cis‑acting elements analyses
The co-expression network analyses of maize SnRK1 genes were constructed using the MaizeNet web server (http://www.inetbio.org/maizenet). MaizeNet is a genome-scale co-functional network of maize genes (Lee et al. 2019). A 2000 bp upstream sequence from the ATG start codon, obtained from the Phytozome database v13, was used as the putative promoter region. Cis-acting regulatory elements (CAREs) were examined in the putative promoter sequences using the PLACE database v30 (https://www.dna.affrc.go.jp/PLACE/?action=newplace) (Higo et al. 1999).
Protein modeling and topology analyses
The putative 3D models of the SnRK1 were generated using the Phyre2 (protein homology/analogy recognition engine v2.0) web tool (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index), which uses advanced remote homology recognition methods to generate 3D models at intensive mode (Kelley et al. 2015). The topological properties were evaluated using the CASTp v3.0 web server (http://sts.bioe.uic.edu/castp/calculation.html) (Tian et al. 2018).
Plant materials and stress treatments
F1 seeds of the maize (Zea mays) cultivar ‘Albayrak’ (Tarex Co., Turkiye) were utilized in this study. These seeds were planted individually in 8 × 8 × 9 cm3 pots filled with a peat–perlite mixture (3:1), and grown in a greenhouse under 28 ± 1 °C, 12:12-h photoperiod without the use of any insecticides. The plants were irrigated with tap water every second day.
For the induction of drought stress, 21-day-old plants were subjected to withholding of water supply for a duration of 7 days. On the other hand, control plants received irrigation according to the regular schedule, i.e., every other day. After the 7-day water withholding period, all plant leaves and roots were collected and preserved at −80 °C for subsequent analyses.
Cold stress was imposed by subjecting the plants to a temperature of + 4 °C for 24 h. Meanwhile, control plants were maintained in the greenhouse under standard conditions. After the cold exposure, all plant leaves and roots were harvested for RNA isolation.
The population of corn leaf aphid (Rhopalosiphum maidis) employed in the experiments was cultivated on maize plants in growth chambers at a temperature of 26 ± 1 °C, relative humidity of 60 ± 10% in 16:8-h photoperiod within plexiglass cages. Weekly checks were conducted on the plants inside the cages, which were watered every other day. Additionally, once a week, dried plants were replaced with fresh ones. A total of 15 aphids were placed on the second true leaf (V2) of the 3-week-old plant (Pingault et al. 2021; Tzin et al. 2015). The plants were exposed to aphid feeding for 2, 4 or 8 h, and the V2 leaves and roots were harvested for RNA isolation at each time point.
RNA isolation and gene expression analysis
Total RNA was isolated from leaf and root tissues using the NucleoZOL kit (Macherey–Nagel, Germany) following manufacturer’s guidelines. Subsequently, Turbo DNase (Thermo Fisher, USA) treatment was performed on the samples. Evaluation of RNA integrity and potential DNA contamination was done through gel electrophoresis, and quantification was carried out using the Qubit system (Invitrogen, USA). Real time-quantitative PCR (RT-qPCR) was conducted on Rotor-Gene Q (Qiagen, USA) using 10 ng DNase-treated RNA with Luna Universal One-Step RT-qPCR Kit (NEB, USA). The primers for RT-qPCR analysis are given in Table 1. The MEP (Membrane protein PB1A10.07c) gene served as an endogenous control (Manoli et al. 2012) in the gene expression analysis using the ΔΔCT method (Livak and Schmittgen 2001), in which the average CT values were derived from a minimum of four biological and three technical replicates for each ZmSnRK1 gene.
MDA (Malondialdehyde) and H2O2 assays
The MDA content of plants was assayed using a method modified from Ohkawa et al. (1979). Grounded in liquid nitrogen, 0.2 g of leaf tissues were suspended in 2 ml of 5% trichloroacetic acid (TCA). The homogenates were transferred into clean 2 ml microfuge tubes and centrifuged at 12,000 rpm at room temperature. Equal amounts of lysate and freshly prepared 0.5% thiobarbituric acid (TBA) in 20% TCA were mixed, then incubated at 96 °C for 25 min. The tubes were chilled on ice until they reached room temperature, then centrifuged at 10,000 rpm for 5 min. The absorbances of the supernatants were measured at 532 nm and 600 nm wavelengths to eliminate non-specific reflections due to turbidity. A freshly prepared 0.5% TBA in 20% TCA solution was used as a blank. The MDA content of the samples was assayed using an absorbance coefficient of 155 mM−1 cm−1.
The H2O2 (hydrogen peroxide) contents were assayed using the method developed by Sergiev et al. (1997). After grounding in liquid nitrogen, 0.5 g of a plant sample was homogenized in 5 ml of 0.1% trichloroacetic acid (TCA). Following centrifugation at 12,000 g for 15 min, 0.5 ml of supernatants were transferred into clean 1.5 ml microfuge tubes, and 0.5 ml of 10 mM potassium phosphate buffer (pH 7.0) and 1 ml of 1 M potassium iodide (KI) were added. The absorbances of the supernatants were measured at 390 nm. The H2O2 contents of the samples were calculated using a standard curve prepared according to known H2O2 concentrations.
Results
Identification of ZmSnRK1 genes and sequence analysis
In this study, four putative SnRK1 genes were identified in the maize genome (Table 2). The number of exons in the SnRK1 genes varied between 10 and 12, and the protein length ranged from 375 to 653 amino acid residues. The molecular weight of the SnRK1 proteins ranged from 42.61 to 72.28 kDa, and the pI values ranged from 7.02 to 9.09. Subcellular localization analyses predicted that SnRK1 proteins are generally found in the cytoplasm.
Domain analysis using the InterPro database revealed the presence of the conserved AMPK/SnRK1 domains in the ZmSnRK1 proteins (Fig. 1): the catalytic domain of the α subunit of the Ser/Thr Kinase (cd14079), the C-terminal regulatory domain of the 5′-AMP-activated protein kinase (AMPK) α catalytic subunit (cd12122), the UBA domain found in the plant sucrose nonfermenting-1-related kinase (SnRK1) proteins (cd14335), and the protein kinase domain profile (PS50011).
Further, ZmSnRK1 proteins were found to have 69.60–82.20% identify with the Arabidopsis SnRK1 orthologs, KIN1 and KIN2, and 73.36–90.03% identify with the rice SnRK1A and SnRK1B. As expected, the similarity ratios of ZmSnRK1 proteins with the rice SnRK1 orthologs were higher than with the Arabidopsis SnRK1 orthologs (Table 3).
To get more insights about ZmSnRK1 gene diversity level, nucleotide pairwise distance analyses were performed, and values were found to range from 0.217 to 0.447 with an overall average of 0.319. The number of segregating sites (S) and nucleotide diversity were found to be 651 and 0.134, respectively. The Tajima test statistic was also found to be −1.45.
Phylogenetic and conserved motif analyses
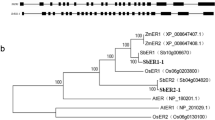
The phylogenetic tree topology clearly showed the separation of SnRK1 proteins in monocots and dicots (groups A and B, respectively). When the monocot–dicot phylogenetic groups were analyzed, it was found that monocots, especially, grouped with stronger bootstrap values (Fig. 2A). Within group A, maize and rice SnRK1 proteins were found to group together (OsSnRK1B and ZmSnRK1.1) with a 100% bootstrap value. Additionally, OsSnRK1A and ZmSnRK1.4 proteins were found to group with a 99% bootstrap value. These results support the strong conservation of SnRK1 genes, especially among monocots. A total of seven different plant species, five dicots and two monocots, were analyzed, and 10 conserved motifs were found (Fig. 2B), namely motif 1 through 10: DGHFLKTSCGSPNYAAPEVISGKLYAGPEVDVWSCGVILYALLCGTLPFD (motif 1), DIYVVMEYVKSGELFDYIVEKGRLQEDEARRFFQQIISGVEYCHRNMVVH (motif 2), IPNLFKKIKGGIYTLPSHLSPGARDLIPRMLVVDPMKRITIPEIRZHPWF (motif 3), QFPVERKWALGLQSRAHPREIMTEVLKALQELNVCWKKIGHYNMKCRWVP (motif 4), SFGKVKIAEHILTGHKVAIKILNRRKIKNMEMEEKVRREIKILRLFMHPH (motif 5), YLAVPPPDTAQQAKKIDEEILQEVVKMGFDKNQLIESLRNRLQNEATVAY (motif 6), KSPNVVKFEIQLYKTRDEKYLLDLQRVQGPQLLFLDLCAAFLTQLRVL (motif 7), PENLLLDSKCNVKIADFGLSN (motif 8), YLLLDNRFRATSGYLGAEFQESMESSFNQIAS (motif 9), and GAGRVENPLPNYKJGKTLGIG (motif 10). Except for the ZmSnRK1.1 and ZmSnRK1.3 proteins, these 10 motifs were detected in all monocot and dicot SnRK1 proteins.
AMPK, SNF1 and SnRK1 undergo activation through T-loop phosphorylation at Thr residue, mediated by upstream kinases (Crozet et al. 2014). The presence of the T-loop motif (LKTS) was confirmed through multiple sequence alignment of ZmSnRK1 genes. In rice, the T-loop is positioned at Thr173; in wheat, at Thr170; and in Arabidopsis, at Thr175. Similarly, the position of the T-loop was discerned at Thr170 and Thr173 in ZmSnRK1.2 and ZmSnRK1.4, respectively, and at Thr234 and Thr383 in ZmSnRK1.1 and ZmSnRK1.2, respectively.
Cis-element analysis of the ZmSnRK1 promoter regions in maize
To understand the regulation of the four ZmSnRK1 genes, 2000 bp upstream regions of each gene were analyzed for the presence of stress-related cis-elements using the PLACE database (Table 4). Of the 10 different stress-related cis-elements examined by the program, only the SARE (salicylic acid responsive cis-acting element) motif (CC(A/T)6GG) was absent in the four ZmSnRK1 gene promoter regions. The rest were all found at different frequency in one or more ZmSnRK1 gene. The most found cis-element was the MYC motif (CACGTG) with 56 occurrences, followed by the W-box (TTGACY) (52), the CG-box (ACCGCC or GCCGAC) (25) and the ABRE (ACGTGG/TC) (24). When comparing the total number of cis-elements in the promoter regions of each gene, the highest number was found in the ZmSnRK1.1 with 58 cis-elements, followed by ZmSnRK1.3 with 52, ZmSnRK1.2 with 49, and ZmSnRK1.4 with 37 cis-elements.
Co-expression network analysis of ZmSnRK1 genes
Gene co-expression network analysis on the MaizeNet web server revealed 440 genes directly linked to ZmSnRK1.1 (Zm00001eb013270), ZmSnRK1.2 (Zm00001eb094400), and ZmSnRK1.3 (Zm00001eb293240) (Fig. 3). The database did not contain ZmSnRK1.4; therefore, this analysis focused only on the three ZmSnRK1 genes. The 10 genes with the highest score are as follows: GRMZM5G845175 (Zm00001d025300), GRMZM2G041312 (Zm00001d003864), GRMZM2G064725 (Zm00001d047594), GRMZM2G138814 (Zm00001d028946), GRMZM2G027632 (Zm00001d041849), GRMZM2G130950 (Zm00001d022545), GRMZM2G047774 (Zm00001d034896), GRMZM2G014170 (Zm00001d012817), GRMZM2G135073 (Zm00001d052051), and GRMZM2G143213 (Zm00001d048497).
In particular, the following four genes showed the highest score in the network: AtSNF4, a homolog of yeast sucrose nonfermenting 4; 5′-AMP-activated protein kinase beta-2 subunit; A-type cyclin-dependent kinase; and protein kinase superfamily protein (Table 5). Therefore, ZmSnRK1 genes were generally associated with SnRK, cyclin-dependent kinase and protein kinase.
Predicted 3D structure of ZmSnRK1 proteins
The putative 3D models and charges of ZmSnRK1 proteins were modelled using the Phyre2 server (Fig. 4). These predicted 3D models showed that the topology of protein structures and charges differed between the four ZmSnRK1 proteins, which may be related to their functional diversity. The percentage of structural overlap of the predicted 3D models ranged from 22.28 to 84.17% for the four ZmSnRK1 proteins. The lowest percentage of overlap was found between ZmSnRK1 and ZmSnRK3 (22.28%) and the highest between ZnSnRK2 and ZmSnRK4 (84.17%). Analyses of the surface topography revealed variations in the surface pocket areas, which may be related to substrate binding, and therefore, a specific function of the protein.
The predicted 3D structures of ZmSnRK1 proteins from the Phyre2 web portal (pictures at the top). The color indicates the charge (isoelectric point), where red is positive, blue is negative and white is neutral. The images below show the surface topography of the same proteins using the CASTp 3.0 server. Red areas on the protein models indicate surface pockets
Morphological and physiological analyses
To evaluate the impact of aphid feeding, drought and cold stress on the maize plants, we employed both morphological observations and physiological assays. Following a 7-day period of drought stress, visible wilting occurred due to water deficit (Fig. 5a). Likewise, exposure to cold temperature for 24 h led to the loss of turgor in leaves and wilting of older leaves (Fig. 5b). However, short-term exposure to aphid feeding, lasting a maximum of 8 h, did not result in discernible morphological changes in the plants (Fig. 5c).
MDA formation, a result of lipid membrane peroxidation by ROS, serves as a marker for stress-induced cellular damage (Łukasik and Goławska 2021; Sun et al. 2022; Turk et al. 2020). H2O2, a byproduct of plant aerobic metabolism, is another commonly utilized indicator of oxidative stress. Elevated levels of H2O2 have been noted in plants subjected to aphid feeding, drought, and cold stress, including maize (Hussain et al. 2020; Pant and Huang 2021; Ramazan et al. 2021). Thus, assessing the levels of MDA and H2O2 in plants confirms the effect of the stress at physiological level (Akbudak et al. 2020; 2022).
The accumulation of MDA and H2O2 in leaf tissues of drought-stressed plants indicates considerable impact of drought on maize plants (Fig. 6). In contrast, there was no significant change in MDA and H2O2 contents in the leaves of plants exposed to aphid feeding or cold stress (Fig. 6a, b). Therefore, physiological effects of the short duration of aphid feeding or cold stress were not detected by these markers, but the lengthy period of drought stress resulted in the production of MDA and H2O2, indicating lipid membrane peroxidation by ROS.
Expression profiles of ZmSnRK1 genes under aphid feeding, drought and cold stress
A differential pattern of expression was found for ZmSnRK1.1, ZmSnRK1.2, ZmSnRK1.3 and ZmSnRK1.4 under aphid feeding, drought or cold stress (Fig. 7).
Expression profiles of ZmSnRK1 genes in maize leaves and roots under aphid feeding, drought and cold stress. Control represents the expression of each gene in untreated maize plants. Histograms represent means of four biological and three technical replicates. The error bars indicate the standard error (sdom; n = 3)
The aphid feeding resulted in the upregulation of ZmSnRK1.3 in the leaves, which increased approximately 15-fold within 2 h of stress compared to the control. After 4 h of aphid feeding, a slight upregulation was also observed in the expression of ZmSnRK1.1, ZmSnRK1.2, and ZmSnRK1.4 in leaves, although their expression levels returned to baseline after 8 h. While the expression levels of ZmSnRK1 genes in roots generally exhibited minimal variation compared to control plants under aphid feeding, a noteworthy upregulation of the ZmSnRK1.3 gene was observed, showing an increase of up to threefold after four hours of exposure.
Drought stress predominantly affected the expression of ZmSnRK1.2, which exhibited a 5.5-fold upregulation in leaves compared to the control, while the expression of other ZmSnRK1 genes fluctuated within a narrow range (0.81–1.71). Drought stress prompted the upregulation of ZmSnRK1.1 and ZmSnRK1.2 genes in roots by up to 1.76-fold, concurrently with a significant downregulation of ZmSnRK1.3 and a slight decrease in ZmSnRK1.4 expression.
Cold stress had a minimal impact on the expression of ZmSnRK1 genes in leaves, except for ZmSnRK1.3, which demonstrated a remarkable 11-fold increase compared to control plants. Cold stress similarly induced a significant 23-fold upregulation in the expression of the ZmSnRK1.3 gene in maize roots, while the other three ZmSnRK1 genes experienced downregulation. Notably, ZmSnRK1.3 emerged as the primary gene significantly upregulated in both maize leaves and roots under cold stress, delineating its pivotal role in maize’s response to aphid feeding and cold stress across different plant tissues. However, its involvement in drought stress appeared less pronounced.
Discussion
This study identified four paralogs of SnRK1 α-subunit genes in the maize genome, referred to as ZmSnRK1 that contain the canonical catalytic domain of the Ser/Thr kinase, the UBA domain, and the C-terminal domain. Additionally, T-loop motif in each of the four ZmSnRK1 was identified. These domains are conserved in SnRK1α genes across yeast, human and plants (Jamsheer et al. 2021), and phosphorylation of Thr in the T-loop is critical in the activation of SnRK1 (Crozet et al. 2016). Phylogenetic analysis showed ZmSnRK1 grouped with rice and diverged from Arabidopsis, indicating dicot and monocot separation of the protein sequences. The genes identified in this study align with the maize SnRK1 genes found by Feng et al. (2022). Nonetheless, the predicted subcellular localization differs from that in our analysis. Feng et al. (2022) reported nuclear localization of ZmSnRK1.1 and mitochondrial localization for ZmSnRK1.3 and ZmSnRK1.4. Whereas our study found cytoplasmic or plasma membrane localization for ZmSnRK1s. Similarly, in Arabidopsis, SnRK1 proteins are localized in the cytoplasm or plasma membrane. However, nuclear localization and possibly also mitochondrial localization, occurs in specific tissues under specific conditions (Bitrián et al. 2011). Thus, while subcellular targeting of SnRK1 is certainly possible, it appears to be highly regulated by the environmental signals.
Co-expression analysis of ZmSnRK1 genes found a strong hit with the homologs of the regulatory subunits: Arabidopsis β-subunit and the βγ-subunit also called AtSNF4. These regulatory subunits interact with the α-subunit to form the heterotrimeric AMPK/SNF1/SnRK1 complex. Next, ZmSnRK1 is co-expressed with the homolog of Arabidopsis A-type cyclin dependent kinase (CDKA) that regulates cell cycle progression. While SnRK1-CDKA interaction has not been reported thus far, it is fair to assume that SnRK1 regulates cell cycle progression to allow stress adaptation. Notably, Arabidopsis SnRK1, KIN10, interacts with CDKE1 in the nucleus, and this complex is possibly involved in retrograde signaling (Ng et al. 2013).
Initially identified as a sensor of low nutrients, SnRK1 is also involved in stress signaling by regulating a common set of stress-responsive genes (Rodrigues et al. 2013). Accordingly, several cis-elements that serve as the binding sites for the well-known transcription factors were identified in 5′-region of ZmSnRK1 genes. The most abundant cis-element was the MYC-binding site with 56 occurrences in the four ZmSnRK1 genes. MYC2 is the most characterized plant MYC TF that regulates plant development and stress response through jasmonate signaling (Song et al. 2022; Thompson and Goggin 2006), and the maize homolog of MYC2, ZmMYC7 was found to be involved in the resistance to pathogenic fungi (Cao et al. 2023). Since plant response to aphid feeding is also regulated by JA signaling (Morkunas et al. 2011), ZmSnRK1 could play a broader role in regulating biotic stress responses. Similarly, W-box and ABRE occur frequently in ZmSnRK1 genes, indicating their role in regulating ABA signaling and WRKY mediated stress pathways, respectively. Overall, ZmSnRK1 gene expression is controlled by numerous transcription factors, which in turn regulate developmental and stress-induced pathways.
We analyzed ZmSnRK1 gene expression in response to aphid feeding, drought or cold treatments. Interestingly, ZmSnRK1.3 was upregulated during aphid feeding that contained 21 W-boxes. It is tempting to speculate that this upregulation occurs through WRKY TFs that are strongly induced by aphid feeding (Annacondia et al. 2021; Gao et al. 2010; Kuśnierczyk et al. 2008). Accordingly, ZmSnRK1.3 was upregulated throughout the duration of the experiment (up to 8 h). A stochastic upregulation was also observed in the roots at 4 h post feeding. Further, ZmSnRK1.1 and ZmSnRK1.2 were upregulated in the leaves and roots in the drought condition and ZmSnRK1.3 was strongly upregulated under cold condition in both roots and leaves. Drought and cold treatments broadly encompass dehydration stress that are regulated by DREB TFs (Yamaguchi-Shinozaki and Shinozaki 2006). However, recent transcriptomic analyses found important roles of WRKY and MYC TFs in regulating dehydration stress (Berchembrock et al. 2022; Singh and Laxmi 2015). Thus, differential expression of ZmSnRK1 genes under drought or cold treatments is likely determined by the cis-elements and their interaction with the TFs. In summary, ZmSnRK1 genes represent the a-subunits of the evolutionarily conserved AMPK/SNF1/SnRK1 family of protein kinases that interact with regulatory subunits to form the SnRK1 complex. Like Arabidopsis and rice SnRK1s, ZmSnRK1 appears to play a central role in energy sensing and the response to environmental signals such as abiotic and biotic stress.
Conclusion
SnRK1 genes play crucial roles in signaling pathways, including responses to both biotic and abiotic stresses in plants. This present study analyzed SnRK1 gene family in Zea mays, covering phylogenetic relationships, gene structures, protein motifs and promoter cis-elements. Four ZmSnRK1 genes were characterized, and the dynamic responses of ZmSnRK1 genes have been elucidated through expression profiling under diverse stress conditions such as aphid feeding, drought and cold stress. Our findings contribute to understanding the subcellular localization of ZmSnRK1 proteins, despite discrepancies with previous research regarding their localization. Notably, we observed significant upregulation of ZmSnRK1.3 under aphid feeding and cold stress and ZmSnRK1.2 under drought stress, shedding light on their potential roles in stress response mechanisms. These results underscore the importance of further investigating the functional roles of ZmSnRK1 genes, laying a foundation for future research in plant stress physiology and biotechnology.
References
Akbudak MA, Yildiz S, Filiz E (2020) Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): bioinformatics analyses and expression profiles in response to drought stress. Genomics 112:4089–4099
Akbudak MA, Filiz E, Çetin D (2022) Genome-wide identification and characterization of high-affinity nitrate transporter 2 (NRT2) gene family in tomato (Solanum lycopersicum) and their transcriptional responses to drought and salinity stresses. J Plant Physiol 272:153684
Alderson A, Sabelli PA, Dickinson JR, Cole D, Richardson M, Kreis M, Shewry PR, Halford NG (1991) Complementation of snf1, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proc Natl Acad Sci 88:8602–8605
Annacondia ML, Markovic D, Reig-Valiente JL, Scaltsoyiannes V, Pieterse CMJ, Ninkovic V, Slotkin RK, Martinez G (2021) Aphid feeding induces the relaxation of epigenetic control and the associated regulation of the defense response in Arabidopsis. New Phytol 230:1185–1200
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Berchembrock YV, Pathak B, Maurya C, Botelho FBS, Srivastava V (2022) Phenotypic and transcriptomic analysis reveals early stress responses in transgenic rice expressing Arabidopsis DREB1a. Plant Direct 6:e456
Bitrián M, Roodbarkelari F, Horváth M, Koncz C (2011) BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J 65:829–842
Broeckx T, Hulsmans S, Rolland F (2016) The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot 67:6215–6252
Cao H, Zhang K, Li W, Pang X, Liu P, Si H, Zang J, Xing J, Dong J (2023) ZmMYC7 directly regulates ZmERF147 to increase maize resistance to Fusarium graminearum. Crop J 11:79–88
Carlson M, Osmond BC, Botstein D (1981) Mutants of yeast defective in sucrose utilization. Genetics 98:25–40
Celenza JL, Carlson M (1986) A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233:1175–1180
Chen L, Su SZ, Huang L, Xia F, Qi H, Xie L, Xiao S, Chen Q (2017) The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front Plant Sci 8:1201
Cohen P (1988) Review lecture: protein phosphorylation and hormone action. Proc Biol Sci 234:115–144
Crozet P, Margalha L, Butowt R, Fernandes N, Elias CA, Orosa B, Tomanov K, Teige M et al (2016) SUMO ylation represses Sn RK 1 signaling in Arabidopsis. Plant J 85:120–133
Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Elias CA, Baena-González E (2014) Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci 5:83320
Feng X, Meng Q, Zeng J, Ma W, Liu W (2022) Genome-wide identification of sucrose non-fermenting-1-related protein kinase genes in maize and their responses to abiotic stresses. Front Plant Sci 13:1087839
Gao LL, Kamphuis LG, Kakar K, Edwards OR, Udvardi MK, Singh KB (2010) Identification of potential early regulators of aphid resistance in Medicago truncatula via transcription factor expression profiling. New Phytol 186:980–994
Gasteiger E, Hoogland C, Gattiker A, Se D, Wilkins MR, Appel RD, Bairoch A (2005) Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM (ed) The Proteomics Protocols Handbook, vol. Humana Press, Totowa NJ, pp 571–607
Halford NG, Grahame Hardie D (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol 37:735–748
Halford NG, Hey SJ (2009) Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem J 419:247–259
Hardie DG, Ross FA, Hawley SA (2012a) AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol 19:1222–1236
Hardie DG, Ross FA, Hawley SA (2012b) AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 13:251–262
Hedbacker K, Carlson M (2008) SNF1/AMPK pathways in yeast. Front Biosci 13:2408
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587
Hrabak EM, Chan CWM, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S et al (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132:666–680
Hulsmans S, Rodriguez M, De Coninck B, Rolland F (2016) The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci 21:648–661
Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225–236
Hussain HA, Men S, Hussain S, Zhang Q, Ashraf U, Anjum SA, Ali I, Wang L (2020) Maize tolerance against drought and chilling stresses varied with root morphology and antioxidative defense system. Plants 9:720
Jamsheer KM, Kumar M, Srivastava V (2021) SNF1-related protein kinase 1: the many-faced signaling hub regulating developmental plasticity in plants. J Exp Bot 72:6042–6065
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Kuśnierczyk A, Winge PER, Jørstad TS, Troczyńska J, Rossiter JT, Bones AM (2008) Towards global understanding of plant defence against aphids–timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ 31:1097–1115
Lastdrager J, Hanson J, Smeekens S (2014) Sugar signals and the control of plant growth and development. J Exp Bot 65:799–807
Lee T, Lee S, Yang S, Lee I (2019) MaizeNet: a co-functional network for network-assisted systems genetics in Zea mays. Plant J 99:571–582
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Łukasik I, Goławska S (2021) Biochemical markers of oxidative stress in maize seedlings exposed to rose-grass aphid, Metopolophium dirhodum. Allelopathy J 53:23–34
Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169:807–815
Morkunas I, Mai VC, Gabryś B (2011) Phytohormonal signaling in plant responses to aphid feeding. Acta Physiol Plant 33:2057–2073
Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C et al (2013) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288:3449–3459
Ohkawa H, Ohishi N, Yagi K (1979) Assay for Lipid Peroxides in Animal-Tissues by Thiobarbituric Acid Reaction. Anal Biochem 95(2):351–358
Pant S, Huang Y (2021) Elevated production of reactive oxygen species is related to host plant resistance to sugarcane aphid in sorghum. Plant Signal Behav 16:1849523
Pathak B, Maurya C, Faria MC, Alizada Z, Nandy S, Zhao S, Jamsheer KM, Srivastava V (2022) Targeting TOR and SnRK1 genes in rice with CRISPR/Cas9. Plants 11(11):1453
Pingault L, Varsani S, Palmer N, Ray S, Williams WP, Luthe DS, Ali JG, Sarath G, Louis J (2021) Transcriptomic and volatile signatures associated with maize defense against corn leaf aphid. BMC Plant Biol 21:1–15
Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12:20–28
Ramazan S, Qazi HA, Dar ZA, John R (2021) Low temperature elicits differential biochemical and antioxidant responses in maize (Zea mays) genotypes with different susceptibility to low temperature stress. Physiol Mol Biol Plants 27:1395–1412
Rodrigues A, Adamo M, Crozet P, Margalha L, Confraria A, Martinho C, Elias A, Rabissi A et al (2013) ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25:3871–3884
Rodriguez M, Parola R, Andreola S, Pereyra C, Martínez-Noël G (2019) TOR and SnRK1 signaling pathways in plant response to abiotic stresses: do they always act according to the “yin-yang” model? Plant Sci 288:110220
Sergiev I, Alexieva V, Karanov E (1997) Effect of spermine, atrazine and combination between them on some endogenous protective systems and stress markers in plants. Compt Rend Acad Bulg Sci 51(3):121–124
Singh D, Laxmi A (2015) Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front Plant Sci 6:165462
Smeekens S, Ma J, Hanson J, Rolland F (2010) Sugar signals and molecular networks controlling plant growth. Curr Opin Plant Biol 13:273–278
Song C, Cao Y, Dai J, Li G, Manzoor MA, Chen C, Deng H (2022) The multifaceted roles of MYC2 in plants: toward transcriptional reprogramming and stress tolerance by jasmonate signaling. Front Plant Sci 13:868874
Sun S, Yao X, Liu X, Qiao Z, Liu Y, Li X, Jiang X (2022) Brassinolide can improve drought tolerance of maize seedlings under drought stress: By inducing the photosynthetic performance, antioxidant capacity and ZmMYB gene expression of maize seedlings. J Soil Sci Plant Nutr 22:2092–2104
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027
Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57:755–766
Tian W, Chen C, Lei X, Zhao J, Liang J (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46:W363–W367
Turk H, Erdal S, Dumlupinar R (2020) Carnitine-induced physio-biochemical and molecular alterations in maize seedlings in response to cold stress. Arch Agron Soil Sci 66:925–941
Tzin V, Fernandez-Pozo N, Richter A, Schmelz EA, Schoettner M, Schäfer M, Ahern KR, Meihls LN et al (2015) Dynamic maize responses to aphid feeding are revealed by a time series of transcriptomic and metabolomic assays. Plant Physiol 169:1727–1743
Wurzinger B, Nukarinen E, Nägele T, Weckwerth W, Teige M (2018) The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol 176:1085–1094
Xiong Y, Sheen J (2015) Novel links in the plant TOR kinase signaling network. Curr Opin Plant Biol 28:83–91
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Acknowledgements
This study was supported by Akdeniz University Scientific Research Projects Coordination Unit competitive grant # FBA-2023-6290 to MAA. MAA is thankful for the financial support provided by the Fulbright Visiting Scholar grant from the Turkish Fulbright Commission.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK). Funding was provided by Akdeniz Üniversitesi (Grant number: FBA-2023-6290).
Author information
Authors and Affiliations
Contributions
MAA: Conceptualization, Writing—Original Draft, Writing—Review and Editing, Supervision, Project administration, Funding acquisition. KY and DC: Investigation. EF: Investigation, Formal Analysis. UY: Resources, Validation. VS: Data curation, Writing—Review and Editing.
Corresponding authors
Ethics declarations
Conflict of interest
M. A. Akbudak, K. Yildiz, D. Cetin, U. Yukselbaba, E. Filiz and V. Srivastava declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Akbudak, M.A., Yildiz, K., Cetin, D. et al. Characterization of ZmSnRK1 genes and their response to aphid feeding, drought and cold stress. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-02006-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-02006-2