Abstract
After the last glacial, the Carpathian Basin was repopulated from either eastward or northward colonisation routes for various species; one of these was the emblematic member of the European megafauna, the red deer, Cervus elaphus. We analysed 303 red deer individuals from the middle of the region, in seven Hungarian game reserves, at ten microsatellite loci (C01, C229, T26, T108, T123, T156, T172, T193, T501, T507), to investigate the genetic diversity of these subpopulations. We discovered high levels of genetic diversity of red deer subpopulations; allelic richness values ranging 4.99–7.01, observed heterozygosity 0.729–0.800, polymorphic information content 0.722–0.806, and Shannon’s information index 1.668–2.064. Multi-locus analyses indicated population admixtures of various degrees that corresponded to geographical location, and complex genetic structures were shown by clustering. Populations in the south-western and the north-eastern parts of the region formed two highly separated groups, and the red deer from populations in between them were highly admixed (in western Pannonia/Transdanubia, where the Danube flows into the Carpathian Basin). This pattern corresponds to the distribution of mitochondrial as well as Y-chromosome lineages. Assignment tests showed that a large fraction of individuals (29.4%) are found outside of their population of origin, indicating that the dispersal of red deer is rather common, which could be expected considering the life course of the species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genetic structure of large mammal species is affected by natural and anthropogenic factors. Anthropogenic impacts like selective hunting, translocations, and habitat destruction/fragmentation can blur natural patterns of genetic diversity and relationships (Frantz et al. 2006; Haanes et al. 2010; Dellicour et al. 2011; Carden et al. 2012; Stanton et al. 2016; Zachos et al. 2016; Galarza et al. 2017; Queirós et al. 2020). Such impacts, particularly in game species, can cause the disintegration of populations into several subpopulations with a more or less pronounced genetic exchange (Hartl et al. 2003; Willems et al. 2016; Iacolina et al. 2019; Mihalik et al. 2020). Knowledge of the genetic diversity of populations and the genetic exchange among neighbouring populations has major importance in identifying and evaluating potential problems, e.g. inbreeding, genetic depletion, and introgression from foreign populations; moreover, it can also provide guidance in determining geographical restrictions in the implementation of wildlife management or conservation programmes.
The red deer (Cervus elaphus L. 1758) is one of the most widespread and valuable European game species (Ludt et al. 2004; Milner et al. 2006). Consequently, its populations have been extensively managed, introduced, restocked, and selectively hunted for centuries or even millennia (Martínez et al. 2002; Haanes et al. 2010; Carden et al. 2012; Rivrud et al. 2013; Queirós et al. 2014; Hoffmann et al. 2016; Stanton et al. 2016; Frantz et al. 2017; Galarza et al. 2017). Furthermore, during the last decades, the keeping of deer in enclosures has spread all over the world, and the species is farmed for venison and antler products (Milner et al. 2006; Wada et al. 2010; Zachos and Hartl 2011; Bana et al. 2018).
Studies on the genetic structure of European mammals, including red deer, based on mitochondrial DNA (mtDNA) sequences (Ludt et al. 2004; Skog et al. 2009; Niedziałkowska et al. 2011; Meiri et al. 2013; Frank et al. 2017; Rey-Iglesia et al. 2017; Queirós et al. 2019) as well as on microsatellites (Zachos et al. 2016; Frantz et al. 2017; Queirós et al. 2019) have shown the large-scale genetic pattern shaped by the Late Pleistocene and Holocene glacial-interglacial cycles. Local or regional red deer populations have also been extensively studied from a population genetic point of view, often taking into account anthropogenic influences (Kuehn et al. 2003; Zachos et al. 2003; Frantz et al. 2006; Hajji et al. 2008; Haanes et al. 2010; Dellicour et al. 2011; Radko et al. 2014; Krojerová-Prokešová et al. 2015; Borowski et al. 2016; Carranza et al. 2016; Hoffmann et al. 2016; Willems et al. 2016; Galarza et al. 2017).
The Carpathian Basin can be considered as a “hotspot” region since it includes crossroads of colonisation routes, where lineages of various species from different refugia are present (Hewitt 2004; Sommer and Zachos 2009). Our previous studies with STRs have shown the existence of two distinct groups of red deer within Hungary (Szabolcsi et al. 2014) and that red deer migrated into the Carpathian Basin from two directions, the west and the south (Frank et al. 2017). The presence of two clades for mitochondrial lineages and Y-chromosomes was also demonstrated (Frank et al. 2017, 2020), and concordant results were also presented by studies on mitochondrial cytochrome-b sequences (Markov et al. 2015).
We hereby present a microsatellite data set of red deer populations covering the centre of the Carpathian Basin (Fig. 1). The main goal of our work was to determine the genetic diversity and genetic structure of populations across this particular geographic region. Furthermore, our study aimed (1) to use microsatellite data to calculate genetic diversity and effective population size in Hungarian red deer; (2) to uncover the nuclear genetic structure of deer populations that provides further insight into the distribution of genetic variability in this particular geographic region; and (3) to search for signatures caused by the post-glacial re-colonisation.
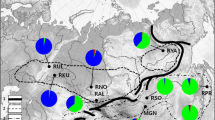
Geographical location of the red deer sampling sites in Hungary. SWG, Gemenc (n = 79); SW2, Baranya (n = 58); SW1, Somogy (n = 56); W1, Zala (n = 27); W2, Csorna (n = 9); NE1, Zemplén (n = 39); NE2, Szabolcs (n = 35). For more details, see Materials and methods
Materials and methods
Sampling
Muscle tissue was obtained from free-ranging adult red deer shot over two consecutive hunting seasons (2014–2016) across Hungary, localised in the centre of the Carpathian Basin (Fig. 1). In total, individual muscle samples of 303 red deer were collected in the course of legally organised hunting events. Animals were not killed for the purposes of this study; thus, no specific ethical approval was needed. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The samples originated from seven sampling sites (hunting reserves) in four regions of the country: Gemenc National Forest (SWG, n = 79), South-Western Hungary (Vajszló, Baranya county, SW2, n = 58; Lábod, Somogy county, SW1, n = 56), Western Hungary (Keszthely, Zala county, W1, n = 27; Csorna, Győr-Moson-Sopron county, W2, n = 9), and North-Eastern Hungary (Zemplén Mountains, NE1, n = 39; Gúth, Szabolcs-Szatmár-Bereg county, NE2, n = 35). The two farthest regions SWG and NE Hungary are separated by historical barriers to gene flow (unbroken flooded lands, sandy hill range, and steppe between the two large rivers, Danube and Tisza, and later, industrialisation and the main rail- and highways). Tissue samples were preserved in 96% ethanol and stored at − 20 °C. Total genomic DNA was extracted using the commercial Genomic DNA Mini Kit (Geneaid Biotech Ltd., New Taipei City, Taiwan) according to the manufacturer’s protocol, and stored at − 20 °C until processing.
Microsatellite genotyping
Samples were genotyped by the DeerPlex system at ten microsatellite (STR) loci (C01, C229, T26, T108, T123, T156, T172, T193, T501, T507) in two multiplex polymerase chain reactions (PCR) using the QIAGEN Multiplex PCR Kit (Qiagen GmbH., Hilden, Germany). Detailed information about the PCR compositions, primer concentrations, and the allelic nomenclature of the STRs can be found in Szabolcsi et al. (2014), with further supplements in Frank et al. (2020). Of the STRs, T507 is X-chromosomal; CO1 and T26 are distantly linked on deer chromosome 14. All DeerPlex STRs were identified in the deer genome sequence CerEla1.0 (Bana et al. 2018).
Amplification reactions were performed using a LifeECO Thermal Cycler (Hangzhou Bioer Technology Co. Ltd., Hangzhou, China); cycling conditions were as described in Szabolcsi et al. (2014), except for the initial denaturation, which was adjusted to 15 min for the multiplex PCR kit. PCR products were separated using an ABI 3100 Genetic Analyser with LIZ500 Size Standard (Applied Biosystems, Foster City, CA, USA), and allele sizes were scored using PeakScanner version 1.0 (Applied Biosystems). Reactions were repeated on samples with incomplete genetic profiles until obtaining a complete ten-locus STR profile.
Multi-locus analysis
Microchecker version 2.2.3 (Van Oosterhout et al. 2004) was employed to search for null alleles, scoring errors, and large allele dropout. To avoid re-sampling of individuals, the identity analysis of the CERVUS version 3.0.6 (Kalinowski et al. 2007) was carried out. The number of alleles per locus (NA), the expected (HE) and observed heterozygosity (HO), deviations from Hardy–Weinberg equilibrium after Bonferroni correction (HWE), and measures of genetic diversity as well as F-statistics (Wright 1951) for each locus and averaged across the 10 loci were calculated with the CERVUS software (Kalinowski et al. 2007) and GenAlEx version 6.501 (Peakall and Smouse 2012).
Effective population sizes (Ne) were calculated using a bias-corrected version of the linkage disequilibrium method by Waples and Do (2008) as implemented in the NEEstimator version 2.1 software (Do et al. 2014). In general, this approach is reliable if effective population sizes are not much larger than 200, and the data set is based on ten or more loci and population sample sizes of 25 or more. These conditions are not met for all populations; therefore, results should be viewed with due caution. Effective population size denotes the number of breeding individuals in an idealised population with the same gene frequency drift or inbreeding as that of the population observed (Wright 1951). More generally, the effective population size may be defined as the number of individuals in an idealised population that have population genetic parameters (i.e. values for HE, HO, PIC, PI, I) identical to those calculated for the actual population investigated. Effective population sizes were calculated for pre-defined populations, not for the clusters retrieved by genetic clustering.
Assessing genetic structure
Several different approaches were used to assess population differentiation (e.g. STRUCTURE, PCoA, DAPC), as suggested previously (Pearse and Crandall 2004). Firstly, the Bayesian clustering method and Markov chain Monte Carlo (MCMC) simulation implemented in STRUCTURE version 2.3.4 (Pritchard et al. 2000) were used to infer the most probable number of genetic clusters without a priori definition of populations. The analyses were run using an admixture model and correlated allele frequencies with a burn-in period of 250, 000 replicates and a sampling period of 750, 000 replicates for the number of clusters (K) from one to six with ten independent runs for each K. To determine the number of genetic clusters, we used the method of Evanno et al. (2005) as implemented by the programme Structure Harvester version 0.6.94 (Earl and VonHoldt 2012).
Principal coordinate analysis (PCoA) was performed on individual multi-locus genotypes by the programme GenAlEx (Peakall and Smouse 2012), to graphically represent the genetic distance matrix between the red deer tested.
Another approach used was a discriminant analysis of principal components (DAPC) implemented in the package adegenet version 2.1.1 (Jombart 2008) that identifies clusters of individuals without using any population genetic model (Jombart et al. 2010). We used the “find.clusters()” function for the identification of the optimal number of clusters (K) based on the Bayesian information criterion (BIC). DAPC was employed to assign individuals into populations, retaining all the principal components, as suggested in the manual. These analyses were run with R version 3.4.4 (R Core Team 2018).
Additionally, the probability of an animal belonging to a population was calculated using the partial Bayesian approach of Rannala and Mountain (1997) implemented in GeneClass version 2.0.h (Piry et al. 2004), with 10, 000 simulated multi-locus genotypes and a threshold for individual exclusion of 0.01, as an exclusion threshold of this magnitude is normally used in ecological studies to identify genetic immigrants (Frantz et al. 2017).
Results
Microsatellite diversity
All analysed loci were highly polymorphic, with the number of alleles per locus ranging from 7 (C229) to 27 (T156), with an average of 17.1. The overall observed and expected heterozygosities stood at 0.767 and 0.834, respectively (Table 1). After Bonferroni correction for multiple tests, only two instances of a locus deviating from Hardy–Weinberg equilibrium (HWE) were observed in one of the pre-defined populations with sample numbers larger than ten (T26 in SW1 and T172 in NE1). No locus significantly deviated from HWE in more than one population, and no locus systematically deviated from HWE. All loci were therefore included in subsequent multi-locus analyses.
Multi-locus analyses
Observed heterozygosity values were high across populations, ranging from 0.729 (NE1) to 0.800 (W1). Polymorphic information content (PIC) ranged between 0.424 (C229) and 0.912 (T193) per locus with a mean value of 0.819. Diversity values were also high in six of seven populations (Table 2). The exception, the values (AR = 4.99; PIC = 0.722; I = 1.668) in the W2 population was expected, due to the low number of individuals. Although the overall FST of 0.048 indicates fairly little genetic differentiation, all but two of the pairwise comparisons were significant after Bonferroni correction (Table 3), suggesting some level of substructuring in Hungarian red deer populations.
The Ne values calculated with the linkage disequilibrium method are given in Table 4. The infinite values for the W2 population may be biased by the small number of samples.
Assessing genetic structure
Genetic structure was detected by STRUCTURE, PCoA, and DAPC analyses. The Bayesian clustering analysis detected a high plateau for the average likelihood scores at around four genetic clusters (K = 4, Fig. 2a), whereas the second-order rate of change in log-likelihood scores showed the highest peak at K = 4 (Fig. 2b). The genotypes were mixed, varied along a wide scale, with Gemenc (SWG) and NE Hungary at the two opposite ends (i.e. with the least mixed genotypes, Fig. 2c).
Bayesian clustering of red deer. a The mean log-likelihood values for each number of clusters. b The probability of the models according to the number of clusters. c The assignment of the samples at K = 2 through K = 5. The genetic groups that were identified at each K value are indicated by distinct colours; each vertical bar within the plots represents an individual red deer grouped according to sampling populations
The PCoA shed light on the differences between the genetic structures of the samples as shown in Fig. 3. The two extreme genetic structures corresponded to the two most separated populations, SWG and the combined NE populations, whereas the combined W populations were in the centre, overlapped toward both the southward and the eastward gene pools, an indication for the mixing zone of the two post-glacial immigration.
The DAPC also revealed the presence of genetic subunits. Comparable BIC values were observed for K = 3 through K = 5, with the lowest BIC obtained for a model postulating four clusters (K = 4, Fig. 3a). Additionally, the assignment of individuals to clusters did not correspond precisely to the geographic origin of samples, an indication for migrations of deer in zones between neighbouring hunting resorts, or by translocations (in next section). In the DAPC analyses, like in the previous STRUCTURE one, samples from SWG and NE Hungary formed the two most distinct clusters (Fig. 3b).
Altogether 70.6% (214/303) of the samples were correctly assigned to their original population using the partial Bayesian approach implemented in GeneClass. An additional 11.6% (35/303) of the individuals were assigned to populations adjacent to their geographical origin. This could indicate that the dispersal of individuals among neighbouring populations is rather common, especially since the number of incorrectly assigned individuals was particularly high in Baranya (SW2) and Somogy (SW1), the two regions that are closest to each other. Seven animals of the 303 samples could not be assigned to any of the sampled populations at the 0.01 threshold level (Table 5).
Discussion
According to the present study, the red deer in the Carpathian Basin appeared to be among the most genetically diverse populations in Europe based on nuclear microsatellite markers. This corresponds to the high genetic diversity of mtDNA haplotypes found previously using cytochrome-b (Markov et al. 2015) and control region sequences (Niedziałkowska et al. 2011; Frank et al. 2017). The high diversity is not surprising, given that the Carpathian Basin is regarded as a contact zone between different red deer lineages (Frank et al. 2017), and populations living in this type of contact or admixture zones can obtain higher genetic diversity than populations living farther away from such zones (Krojerová-Prokešová et al. 2015; Borowski et al. 2016). Our analyses of 303 individual multi-locus genotypes of Hungarian red deer have uncovered a rather high genetic diversity across Hungary. The lowest diversity was found in the Csorna (W2) population in Western Hungary, probably due to the low number of samples analysed. Heterozygosity and genetic diversity values of Hungarian deer were high compared with values previously reported for other European populations (Supplementary Table S1) from western (Dellicour et al. 2011), central, and northern (Kuehn et al. 2003; Kuehn et al. 2004; Krojerová-Prokešová et al. 2015; Radko et al. 2014; Hoffmann et al. 2016; Willems et al. 2016), as well as Mediterranean regions of Europe (Hajji et al. 2008; Karaiskou et al. 2014; Queirós et al. 2014, 2019; Carranza et al. 2016). However, this should be taken with due caution, because diversity estimates might be influenced by differences in sample size, marker number, and sets of markers used, reducing the comparability between studies (Queirós et al. 2015; Reiner et al. 2019).
The overall nuclear genetic structure of red deer in Hungary was more complex than that found in mitochondrial phylogenetic studies (Niedziałkowska et al. 2011; Markov et al. 2015; Frank et al. 2017). Although all clustering methods indicated some structuring, the established clusters greatly overlapped and could not be separated clearly, and they corresponded only approximately to the geographic origin of the samples. Various natural and anthropogenic factors may contribute to the lack of a clear population structure. The natural dispersal of individuals would be sufficient for sustaining the gene flow between populations, thus blurring the genetic substructure (Kuehn et al. 2004; Haanes et al. 2010; Fickel et al. 2012; Carranza et al. 2016; Steinbach et al. 2018). A notable part of the individuals analysed, 11.6% (35/303), were assigned to populations adjacent to their geographical origin. This could indicate that the dispersal of individuals among neighbouring populations can be a common phenomenon or that finely structured populations are hard to distinguish using genetic markers. In natural populations, red deer usually has a male-biased dispersal with females being philopatric (Clutton-Brock and Lonergan 1994; Pérez-Espona et al. 2008; Loe et al. 2009; Jarnemo 2011; Fickel et al. 2012; Kropil et al. 2015), although female deer may also exhibit some migration behaviour (Clutton-Brock and Lonergan 1994; Kuehn et al. 2003, 2004; Pérez-Espona et al. 2008; Borowski et al. 2016). Gene flow by dispersal is likely to have occurred from the re-colonisation of the Carpathian Basin to the present, and due to the relatively small geographic distances between populations, some gene flow among them likely remained to date.
The blurred structuring could also be explained by human-induced translocations, as such activities are thought to have concerned important game species for centuries or even millennia (Scandura et al. 2011; Zachos and Hartl 2011; Steinbach et al. 2018; Queirós et al. 2020). It is believed that the present gene pool of many European red deer populations is affected by human-induced translocations (Frantz et al. 2006; Carden et al. 2012; Krojerová-Prokešová et al. 2015; Stanton et al. 2016; Galarza et al. 2017; Iacolina et al. 2019; Queirós et al. 2020). However, the results of the assignment test, namely, the low number of misassigned deer, indicate that the transportation of deer from greater distances was not common in this region. The possibility of human-induced translocations between nearby populations could not be ruled out, as the effect of such translocations on the genetic structure is similar to the effects of natural dispersal (Kuehn et al. 2003; Frantz et al. 2006, 2017; Zachos et al. 2016).
Our genetic clustering results indicated a highly consequent separation of south-western (SWG) and northern/north-eastern (NE1 and NE2) populations, whereas populations between these regions showed highly mixed genotypes (i.e. SW1, SW2, W1, see Figs. 1, 2c, and 3). This pattern corresponds to the previous description of the presence and distribution of the European red deer mitochondrial lineages (Frank et al. 2017) as well as Y-chromosome lineages of red deer (Frank et al. 2020). Thus, our data strengthen the presumption that the Carpathian Basin represents an admixture zone between southern and western European red deer lineages. Comparing the autosomal markers with previous mitochondrial studies (Markov et al. 2015; Frank et al. 2017), the position of the admixture zone between the lineages corresponds to the land where the River Danube flows into the Carpathian Basin in West-Pannonia (currently named Western Transdanubia). Unfortunately, this admixture zone could not be localised more precisely due to the low number of samples obtained from the region.
The number of individuals excluded from all sampled populations was seven, which corresponds to a frequency of around two per cent. This value is lower than that previously inferred in Western European red deer populations (Frantz et al. 2017), and these animals may have immigrated naturally from neighbouring, not sampled populations. The moderate dispersal of individuals between populations could be expected due to the territory-adherent behaviour of red deer (Clutton-Brock and Lonergan 1994; Kuehn et al. 2003; Pérez-Espona et al. 2008; Loe et al. 2009; Jarnemo 2011; Kropil et al. 2015).
Overall, estimated effective population sizes were in the range of those previously calculated for European red deer based on genetic data (Martínez et al. 2002; Kuehn et al. 2003; Zachos et al. 2016). The smallest value was calculated for the W1 population (NE = 41.6 for the frequency threshold of 0.05) and the largest for Gemenc, SWG (NE = 529.7 for the frequency threshold of 0.05) (Fig. 4). The infinite values for the W2 population are probably due to the low sample size. While the linkage disequilibrium approach is viewed as a reliable method, there are many unknowns in any calculation of effective population size (Luikart et al. 2010). The values therefore might best be viewed in a comparative context rather than as absolute values for each of the populations separately.
A more detailed genetic study will be necessary to get a better insight into the effect of geographic distance and landscape features on the population genetic structure of red deer in the Carpathian Basin. The analysis of a larger number of populations would refine our knowledge about the genetic structuring of the species in Central Europe. Furthermore, an analysis of ancient DNA of red deer samples of the region could provide a deeper investigation of past events and result in a more precise view of recent and historical patterns of demographic changes.
References
Bana ÁN, Nyiri A, Nagy J, Frank K, Nagy T, Stéger V, Schiller M, Lakatos P, Sugár L, Horn P, Barta E, Orosz L (2018) The red deer Cervus elaphus genome CerEla1.0: sequencing, annotating, genes, and chromosomes. Mol Genet Genomics 293:665–684
Borowski Z, Świsłocka M, Matosiuk M, Mirski P, Krysiuk K, Czajkowska M et al (2016) Purifying selection, density blocking and unnoticed mitochondrial DNA diversity in the red deer Cervus Elaphus. PLoS ONE 11:e0163191
Carranza J, Salinas M, de Andrés D, Pérez-González J (2016) Iberian red deer: paraphyletic nature at mtDNA but nuclear markers support its genetic identity. Ecol Evol 6:905–922
Carden RF, McDevitt AD, Zachos FE, Woodman PC, O’Toole P, Rose H et al (2012) Phylogeographic, ancient DNA, fossil and morphometric analyses reveal ancient and modern introductions of a large mammal: the complex case of red deer (Cervus elaphus) in Ireland. Quat Sci Rev 42:74–84
Clutton-Brock TH, Lonergan ME (1994) Culling regimes and sex ratio biases in Highland red deer. J Appl Ecol 31:521–527
Dellicour S, Frantz AC, Colyn M, Bertouille S, Chaumont F, Flamand MC (2011) Population structure and genetic diversity of red deer (Cervus elaphus) in forest fragments in north-western France. Conserv Genet 12:1287–1297
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fickel J, Bubliy OA, Stache A, Noventa A, Jirsa A, Heurich M (2012) Crossing the border? Structure of the red deer (Cervus elaphus) population from the Bavarian-Bohemian forest ecosystem. Mamm Biol 77:211–220
Frank K, Bleier N, Tóth B, Sugár L, Horn P, Barta E, Orosz L, Stéger V (2017) The presence of Balkan and Iberian red deer (Cervus elaphus) mitochondrial DNA lineages in the Carpathian Basin. Mamm Biol 86:48–55
Frank K, Bana ÁN, Bleier N, Sugár L, Nagy J, Wilhelm J, Kálmán Z, Barta E, Orosz L, Horn P, Stéger V (2020) Mining the red deer genome (CerEla1.0) to develop X-and Y-chromosome-linked STR markers. PLoS ONE 15:e0242506
Frantz AC, Tigel Pourtois J, Heuertz M, Schley L, Flamand MC, Krier A et al (2006) Genetic structure and assignment tests demonstrate illegal translocation of red deer (Cervus elaphus) into a continuous population. Mol Ecol 15:3191–3203
Frantz AC, Zachos FE, Bertouille S, Eloy MC, Colyn M, Flamand MC (2017) Using genetic tools to estimate the prevalence of non-native red deer (Cervus elaphus) in a western European population. Ecol Evol 7:7650–7660
Galarza JA, Sánchez-Fernández B, Fandos P, Soriguer R (2017) Intensive management and natural genetic variation in red deer (Cervus elaphus). J Hered 108:496–504
Haanes H, Røed KH, Mysterud A, Langvatn R, Rosef O (2010) Consequences for genetic diversity and population performance of introducing continental red deer into the northern distribution range. Conserv Genet 11:1653–1665
Hajji GM, Charfi-Cheikrouha F, Lorenzini R, Vigne JD, Hartl GB, Zachos FE (2008) Phylogeography and founder effect of the endangered Corsican red deer (Cervus elaphus corsicanus). Biodivers Conserv 17:659–673
Hartl GB, Zachos FE, Nadlinger K (2003) Genetic diversity in European red deer (Cervus elaphus L.): anthropogenic influences on natural populations. C. R. Biol.326, 37–42. https://doi.org/10.1016/S1631-0691/2803/2900025-8
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B 359:183–195
Hoffmann GS, Johannesen J, Griebler EM (2016) Population dynamics of a natural red deer population over 200 years detected via substantial changes of genetic variation. Ecol Evol 6:3146–3153
Iacolina L, Corlatti L, Buzan E, Safner T, Šprem N (2019) Hybridisation in European ungulates: an overview of the current status, causes, and consequences. Mamm Rev 49:45–59
Jarnemo A (2011) Male red deer (Cervus elaphus) dispersal during the breeding season. J Ethol 29:329–336
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Karaiskou N, Tsakogiannis A, Gkagkavouzis K, Papika S, Latsoudis P, Kavakiotis I et al (2014) Greece: a Balkan subrefuge for a remnant red deer (Cervus elaphus) population. J Hered 105:334–344
Krojerová-Prokešová J, Barančeková M, Koubek P (2015) Admixture of eastern and western European red deer lineages as a result of postglacial recolonization of the Czech Republic (Central Europe). J Hered 106:375–385
Kropil R, Smolko P, Garaj P (2015) Home range and migration patterns of male red deer Cervus elaphus in Western Carpathians. Eur J Wildl Res 61:63–72
Kuehn R, Haller H, Schroeder W, Rottmann O (2004) Genetic roots of the red deer (Cervus elaphus) population in Eastern Switzerland. J Hered 95:136–143
Kuehn R, Schroeder W, Pirchner F, Rottmann O (2003) Genetic diversity, gene flow and drift in Bavarian red deer populations (Cervus elaphus). Conserv Genet 4:157–166
Loe LE, Mysterud A, Veiberg V, Langvatn R (2009) Negative density-dependent emigration of males in an increasing red deer population. Proc R Soc B 276:2581–2587
Ludt CJ, Schroeder W, Rottmann O, Kuehn R (2004) Mitochondrial DNA phylogeography of red deer (Cervus elaphus). Mol Phylogenet Evol 31:1064–1083
Luikart G, Ryman N, Tallmon DA, Schwartz MK, Allendorf FW (2010) Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv Genet 11:355–373. https://doi.org/10.1007/s10592-010-0050-7
Markov GG, Kuznetsova MV, Danilkin AA, Kholodova MV, Sugár L, Heltai M (2015) Genetic diversity of the red deer (Cervus elaphus L.) in Hungary revealed by Cytochrome b gene. Acta Zool Bulg 67:11–17
Martínez JG, Carranza J, Fernández-García JL, Sánchez-Prieto CB (2002) Genetic variation of red deer populations under hunter exploitation in southwestern Spain. J Wildl Manage 66:1273–1282
Meiri M, Lister AM, Higham TFG, Stewart JR, Straus LG, Obermaier H et al (2013) Late-glacial recolonization and phylogeography of European red deer (Cervus elaphus L.). Mol Ecol 22:4711–4722
Mihalik B, Frank K, Astuti PK, Szemethy D, Szendrei L, Szemethy L et al (2020) Population genetic structure of the wild boar (Sus scrofa) in the Carpathian Basin. Genes 11:1194
Milner JM, Bonenfant C, Mysterud A, Gaillard JM, Csányi S, Stenseth NC (2006) Temporal and spatial development of red deer harvesting in Europe: biological and cultural factors. J Appl Ecol 43:721–734
Niedziałkowska M, Jędrzejewska B, Honnen AC, Otto T, Sidorovich VE, Perzanowski K et al (2011) Molecular biogeography of red deer Cervus elaphus from eastern Europe: insights from mitochondrial DNA sequences. Acta Theriol 56:1–12
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28:2537–2539
Pearse DE, Crandall KA (2004) Beyond FST: analysis of population genetic data for conservation. Conserv Genet 5:585–602
Pérez-Espona S, Pérez-Barbería FJ, McLeod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol Ecol 17:981–996
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Queirós J, Acevedo P, Santos JPV, Barasona J, Beltran-Beck B, González-Barrio D et al (2019) Red deer in Iberia: Molecular ecological studies in a southern refugium and inferences on European postglacial colonization history. PLoS ONE 14:e0210288
Queirós J, Godinho R, Lopes S, Gortázar C, De La Fuente J, Alves PC (2015) Effect of microsatellite selection on individual and population genetic inferences: an empirical study using cross-specific and species-specific amplifications. Mol Ecol Res 15:747–760
Queirós J, Gortázar C, Alves PC (2020) Deciphering anthropogenic effects on the genetic background of the red deer in the Iberian Peninsula. Front Ecol Evol 8:147
Queirós J, Vicente J, Boadella M, Gortázar C, Alves PC (2014) The impact of management practices and past demographic history on the genetic diversity of red deer (Cervus elaphus): an assessment of population and individual fitness. Biol J Linn Soc 111:209–223
R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/
Radko A, Zalewski D, Rubiś D, Szumiec A (2014) Genetic differentiation among 6 populations of red deer (Cervus elaphus L.) in Poland based on microsatellite DNA polymorphisms. Acta Biol Hung 65:414–427
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201
Reiner G, Lang M, Willems H (2019) Impact of different panels of microsatellite loci, different numbers of loci, sample sizes, and gender ratios on population genetic results in red deer. Eur J Wildl Res 65:25
Rey-Iglesia A, Grandal-d’Anglade A, Campos PF, Hansen AJ (2017) Mitochondrial DNA of pre-last glacial maximum red deer from NW Spain suggests a more complex phylogeographical history for the species. Ecol Evol 7:10690–10700
Rivrud IM, Sonkoly K, Lehoczki R, Csányi S, Storvik GO, Mysterud A (2013) Hunter selection and long-term trend (1881–2008) of red deer trophy sizes in Hungary. J Appl Ecol, 50:168–180. https://doi.org/10.1111/1365-2664.12004
Scandura M, Iacolina L, Apollonio M (2011) Genetic diversity in the European wild boar Sus scrofa: phylogeography, population structure and wild x domestic hybridization. Mamm Rev 41:125–137
Skog A, Zachos FE, Rueness EK, Feulner PGD, Mysterud A, Langvatn R et al (2009) Phylogeography of red deer (Cervus elaphus) in Europe. J Biogeogr 36:66–77
Sommer RS, Zachos FE (2009) Fossil evidence and phylogeography of temperate species: ‘glacial refugia’ and post-glacial recolonization. J Biogeogr 36:2013–2020
Stanton DWG, Mulville JA, Bruford MW (2016) Colonization of the Scottish islands via long-distance Neolithic transport of red deer (Cervus elaphus). Proc R Soc B 283:20160095
Steinbach P, Heddergott M, Weigand H, Weigand AM, Wilwert E, Stubbe M et al (2018) Rare migrants suffice to maintain high genetic diversity in an introduced island population of roe deer (Capreolus capreolus): evidence from molecular data and simulations. Mamm Biol 88:64–71
Szabolcsi Z, Egyed B, Zenke P, Pádár Z, Borsy A, Stéger V, Pásztor E, Csányi S, Buzás Z, Orosz L (2014) Constructing STR multiplexes for individual identification of Hungarian red deer. J Forensic Sci 59:1090–1099
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wada K, Okumura K, Nishibori M, Kikkawa Y, Yokohama M (2010) The complete mitochondrial genome of the domestic red deer (Cervus elaphus) of New Zealand and its phylogenic position within the family Cervidae. Anim Sci J 81:551–557
Waples RS, Do C (2008) ldne: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756
Willems H, Welte J, Hecht W, Reiner G (2016) Temporal variation of the genetic diversity of a German red deer population between 1960 and 2012. Eur J Wildl Res 62:277–284
Wright S (1951) The genetical structure of populations. Ann Eugenics 15:323–354
Zachos FE, Frantz AC, Kuehn R, Bertouille S, Colyn M, Niedziałkowska M et al (2016) Genetic structure and effective population sizes in European red deer (Cervus elaphus) at a continental scale: insights from microsatellite DNA. J Hered 107:318–326
Zachos FE, Hartl GB (2011) Phylogeography, population genetics and conservation of the European red deer Cervus elaphus. Mamm Rev 41:138–150
Zachos F, Hartl GB, Apollonio M, Reutershan T (2003) On the phylogeographic origin of the Corsican red deer (Cervus elaphus corsicanus): evidence from microsatellites and mitochondrial DNA. Mamm Biol 68:284–298
Acknowledgements
We are very grateful to Gemenc Forestry and Game Co. Ltd., Északerdő Forestry PLC, Gergely Fazekas, and all local hunters who participated in the sample collection. This research was supported by the Ministry of Agriculture (grant no. NAIK-MBK/MS71411). K. F. was supported by the New National Excellence Program of the Ministry of Human Capacities (grant no. ÚNKP-17-3-I-KE-24).
Funding
Open access funding provided by Hungarian University of Agriculture and Life Sciences. This research was supported by the Ministry of Agriculture (grant no. NAIK-MBK/MS71411). K. F. was supported by the New National Excellence Program of the Ministry of Human Capacities (grant no. ÚNKP-17–3-I-KE-24).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
No animals were killed specifically for the purposes of this study; thus, no specific ethical approval was needed. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Frank, K., Szepesi, K., Bleier, N. et al. Genetic traces of dispersal and admixture in red deer (Cervus elaphus) populations from the Carpathian Basin. Eur J Wildl Res 68, 55 (2022). https://doi.org/10.1007/s10344-022-01602-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-022-01602-w