Abstract
Viral enteritis is a significant threat to domestic dogs. The two primary pathogens that cause viral enteritis in dogs are canine coronavirus (CCoV) and canine parvovirus (CPV). In this study, we investigated the occurrence of CPV-2, CCoV, and canine circovirus coinfection by characterizing circulating subtypes of CPV-2 in faecal samples from symptomatic dogs admitted to veterinary clinics located in Ankara, Elazığ, Kayseri, and Kocaeli provinces of Turkey, between 2019 and 2022. Virus detection by PCR and RT-PCR revealed that CPV-2 was present in 48 (77.4%) samples, and no other agents were detected. Based on the occurrence of the codon GAT at positions 1276 to 1278 (coding for aspartate at residue 426) of VP2, all CPV-2 isolates were confirmed to be of the CPV-2b subtype. The complete genome sequences of two CPV-2b isolates showed a high degree of similarity to and phylogenetic clustering with Australian and East Asian strains/isolates. The predominant CPV strain circulating in the three different regions of Turkey was found to be a CPV-2b strain containing the amino acid substitutions at Y324I and T440A, which commonly contribute to immune escape. This is the first report of complete genomic analysis of CPV-2 isolates circulating in symptomatic domestic dogs in Turkey. The evolution of CPV-2 has raised questions about the efficacy of current vaccination regimes and highlights the importance of monitoring the emergence and spread of new CPV-2 variants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Viral enteritis is a significant threat to domestic and wild dogs worldwide [1]. Canine parvovirus (CPV) is one of the primary causative agents of gastroenteritis, but survival rates can be as high as 80-95% when the infection treated early and aggressively. CPV-infected dogs without treatment can have survival rates as low as 9.1% [2, 3].
The non-enveloped virions of CPV contain a 5.2-kb long, single-stranded DNA genome with a mutation rate of approximately 10-4 nucleotide substitutions per site per year, which is similar to that of RNA viruses [4]. CPV is believed to have originated from feline parvoviruses or a closely related FPV-like parvovirus in wild carnivores [5]. The CPV-2 DNA genome contains two open reading frames (ORFs) translated into four proteins by alternative splicing. The first of the ORFs encodes two non-structural proteins (NS1 and NS2), while the second encodes two structural proteins (VP1 and VP2). An additional protein, VP3, originates from cleavage of VP2 by host proteases [6]. NS1 is a pleiotropic nuclear phosphoprotein [7], which together with NS2, is responsible for viral replication, DNA packaging, cytotoxicity, and pathogenicity [6, 8]. VP2 is the main capsid protein and antigenic marker, constituting 90% of the viral capsid, and it plays a critical role in determining the tissue tropism and host range of the virus [6, 9, 10]. Mutations and recombination events in the VP2 gene affect host range and receptor binding affinity [11], and therefore, better and more-precise identification of the resulting strains is essential for researchers to understand the relationship of circulating strains to vaccine strains and the evolutionary pattern of CPV strains in the field [12].
Canine parvovirus 2 (CPV-2; also called the "original" CPV-2), belongs to the species Carnivore protoparvovirus 1 (genus Protoparvovirus of the family Parvoviridae), together with feline parvovirus virus (FPV), mink enteritis virus (MEV), and raccoon parvovirus (RPV) [13]. CPV-2 was first identified in the mid-to-late 1970s as a major cause of viral enteritis in young dogs, and it subsequently became endemic in dog populations worldwide. In 1980, a new variant, CPV-2a, emerged and replaced the original CPV-2 variant worldwide. Variant CPV-2a acquired the ability to infect both cats and dogs by acquiring four mutations (L87M, I101T, A300G, D305Y) in the capsid protein (VP2) that enable CPV-2 to bind to the feline transferrin receptor (TfR) [14]. In 1984, another antigenic variant emerged as a new CPV type, designated CPV-2b (N426D), which is currently cocirculating with CPV-2a in varying proportions in different geographic regions [15]. Prospectively in 2000, but retrospectively in 1996 [16], a new CPV-2c variant (D426E) was reported in Europe [16] that rapidly spread to many parts of the world in the subsequent years [17,18,19]. Although initial reports claimed that CPV-2c was of low pathogenicity [20], field observations now indicate that most CPV-2c cases were associated with severe hemorrhagic enteritis, frequently with fatal outcomes. CPV-2c has also been reported to be able to infect CPV-2-vaccinated adults repeatedly and cause disease [21]. More recently, CPV-2a and CPV-2b variants with alanine at aa position 297 have been designated as “new CPV-2a” and “new CPV-2b”, respectively, and this is considered their molecular signature [22, 23].
CPV relies entirely on host cell mechanisms and requires active, rapidly proliferating cells, such as intestinal crypts and lymphoid organs, for its replication [6]. This feature largely explains its pathogenesis and clinical picture. Due to the destruction of intestinal villi, the development of severe protein-losing enteropathy through the gastrointestinal tract in puppies with severe CPV enteritis results in dehydration and hypovolemic shock with fluid loss. Dissolution of the intestinal barrier facilitates the passage of bacteria and/or endotoxins into the bloodstream; however, severe immunosuppression, characterized by lymphopenia and ultimately panleukopenia, accompanies the gastrointestinal manifestations. Therefore, septicemia, endotoxemia, systemic inflammation, coagulation disorders, and septic shock are expected in CPV-infected animals and contribute significantly to disease severity and lethality [2, 24].
Canine coronavirus (CCoV) is a single-stranded RNA virus of the family Coronaviridae (genus Alphacoronavirus) [25]. CCoV replicates in the enterocytes at the top of the villi. As a result, the damaged epithelium is no longer replaced by new enterocytes that develop in the crypt, which can lead to severe hemorrhagic enteritis with possible CPV coinfection [26]. Canine circovirus (CCV) is a non-enveloped virus with a single-strand circular DNA genome (approximately 2 kb) [27], belonging to the family Circoviridae, and is associated with vasculitis, hemorrhagic enteritis, and diarrhea [28, 29].
The aim of this study was to investigate the presence of CPV-2, CCoV, and canine circovirus coinfection and characterize circulating subtypes of CPV-2 in symptomatic dogs admitted to veterinary clinics located in Ankara, Elazığ, Kayseri, and Kocaeli provinces of Turkey between 2019 and 2022.
Materials and methods
Sampling and location
Regardless of age, breed, gender, and vaccination status, dogs that displayed clinical signs of CPV-2 infection, such as diarrhea, vomiting, and dehydration, were included in the study after obtaining the consent of their owners. Information about the dogs was obtained by interviewing the owner. Dogs that were reported to have been vaccinated recently (within the previous 30 days) were excluded from the study to circumvent the effects of vaccine strains. The study population consisted of 62 dogs of 15 different breeds, ranging in age from 1.5 to 60 months. While 20 dogs were reported to have been vaccinated at least once, the remaining animals (n = 40) had not been vaccinated at all, and vaccination data were not available for two dogs. Faecal samples were collected aseptically from the rectum of symptomatic dogs admitted to veterinary clinics in Ankara (n = 7), Elazig (n = 24), Kayseri (n = 27), and Kocaeli (n = 4) provinces, which represent the three geographical regions of Turkey, namely the Marmara region, Central Anatolia, and Eastern Anatolia, from November 2019 to January 2022.
Faecal samples were homogenized (10% w/v) in phosphate-buffered saline (PBS) and centrifuged at 10,000 g for 5 min. The supernatants were recovered and treated with gentamicin sulfate (20 mg/mL).
Nucleic acid extraction and gene amplification (RT-PCR, PCR)
Template DNA and RNA were extracted from the supernatants of faecal samples using a commercial extraction kit (MinElute Virus Spin Kit, QIAGEN, Hilden, Germany). Positive control DNA for CPV-2 was derived from freeze-dried Novibac parvo vaccine (MSD Animal Health, UK). Canine circovirus DNA, previously isolated from the feces of a dog with enteritis and confirmed by sequencing, was obtained from Dr. Turhan Turan of Cumhuriyet University and was used as a positive control for CCV in the PCR step. The RNA template obtained from the tissues of a dog with pantropic canine coronavirus, previously confirmed by RT-PCR, was used as a positive control. In each extraction step, faecal samples of clinically healthy dogs were used as negative controls. A blank reaction consisting of primers and no DNA template was included as a reagent control.
Extracted viral nucleic acids were stored at –20°C and were used in molecular tests within 10 working days.
A partial VP2 fragment of CPV-2 (nucleotides 837-1461) was amplified by PCR using a protocol described previously [30]. The complete genome of CPV-2 was characterized as described by Canuti et al. [31]. Viral RNA was extracted from faecal samples screened for CCoV-I and CCoV-II by reverse transcription PCR (RT-PCR) as described by Pratelli et al. [32]. In addition, CCV PCR screening of the extracted nucleic acid was performed as described previously [33]. The nucleotide sequences of the primers used in the PCR and RT-PCR steps and the sizes of the amplified regions are shown in Table 1.
The PCR-amplified products and a 100-bp DNA ladder (Cleaver Scientific, UK) were subjected to electrophoresis in a 1.5% agarose gel containing ethidium bromide (0.5 μg/ml) in Tris-acetate EDTA (TAE) buffer for 40 min at 120 V and visualized using a UV transilluminator.
Sequencing and phylogeny
PCR products were gel-purified and sequenced by a commercial sequencing service (Macrogen Europe, Amsterdam, The Netherlands). Sequencing was performed by the Sanger dideoxy chain termination method in an ABI Prism 3130 Genetic Analyzer (Applied Biosystems, MA, USA), using a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, MA, USA). Bidirectional nucleotide sequence alignments made using Clustal W software were edited, verified with Basic Local Alignment Search Tool (nih.gov) (BLASTn), and submitted to the GenBank database.
The complete genome sequences of two CPV-2 isolates, excluding the non-translated terminal regions, were determined by the Sanger method to investigate the molecular epidemiology of this virus. Overlapping sequences were used to assemble the full-length sequences of the VP- and NS- encoding genes, which were edited and aligned with sequences of related isolates obtained from the GenBank database, using Clustal W.
A phylogenetic tree was constructed based on the two complete genomic, NS1, and VP2 sequences obtained here and an additional 28 sequences obtained from the GenBank database, which included published sequences from different geographic regions, some of which were found by BLASTn analysis to be very similar to those of the two Turkish isolates.
The sequences were aligned using CLUSTAL W, and this alignment was used to generate phylogenetic trees by the maximum-likelihood method, using Tamura-Nei model [34] with 1000 bootstrap replications in MEGA X [35].
Virus isolation
CPV-2-DNA-positive faecal supernatants were clarified and treated with antibiotics and then inoculated onto freshly trypsinized Madin-Darby canine kidney cells (MDCK, NBL-2, ATCC, USA) and grown in Dulbecco’s modified Eagle medium (DMEM, Sigma, MO, USA) containing 5% fetal bovine serum (FBS, Sigma, MO, USA). The isolates were propagated through serial passages on cell cultures until a cytopathic effect was visible in approximately 80% of the cells. Infected cells were frozen and thawed several times, and the presence of the virus in the harvested culture supernatant was confirmed by PCR.
Statistical analysis
Descriptive statistics and frequency distributions were calculated using chi-square analysis, to determine the association between the putative risk factors and CPV-2 positivity. All statistical associations were considered significant at p < 0.05.
Results
PCR and RT-PCR
An amplicon of 625 bp, corresponding to a portion of the VP2 gene, was obtained from in 48 of 62 (77%) samples. A band of the same size was also obtained from the CPV-2 vaccine used as a positive control. Fragments of 694 and 533 bp, corresponding to the amplicons from the positive controls CCoV and CCV, respectively, were detected, but not in clinical specimens. There was no amplification from the negative controls or the template DNA-PCR control (see Supplementary Table S1 for signalment data of CPV-2-positive dogs).
Sequencing and phylogeny
The VP2 amplicons that were obtained from the 48 faecal swab samples were sequenced, and the deduced amino acid sequences of the VP2 proteins indicated that CPV-2 subtype 2b was predominant. The characteristic GAT codon encoding aspartate at nt position 1276-1278 was used to identify CPV-2b isolates. The distribution of CPV-2 isolates from this study (yellow stars) and those reported previously in Turkey [36,37,38,39,40] are shown in Fig. 1.
Distribution of CPV-2 subtypes identified in this study (yellow stars) and those reported previously in Turkey. Uniform blue shading indicates the 2b subtype of CPV-2, while a red-blue gradient indicates the 2a and 2b subtypes of CPV-2. Hashed gray shading indicates the 2a, 2b, and 2c subtypes. The *2a subtype has been reported previously in Ankara, so it is shown with red and blue gradient.
In the 48 samples, eight partial VP2 sequences were identified and submitted to the GenBank database (OM747852-OM7478529). The original CPV type 2, CPV-2a, and CPV-2c were not found. The partial VP2 sequence of the 48 CPV-2 samples had 97.76-100% nt and 99.52-100% aa identity to each other. An amino acid substitution (V to G) was found at position 484 in only two of the 48 sequences.
Two complete genome sequences obtained in the present study were submitted to the GenBank database and were assigned the accession numbers OM721655 and OM721656. Both were from CPV-2b isolates. CPV-2b-K5-TR is 4450 nt long, and CPV-2b-O1-TR is 4452 nt long, and both encode four proteins: NS1 (2007 nt, 668 aa), NS2 (1970 nt, 165 aa), VP1 (2256 nt, 727 aa), and VP2 (1755 nt, 584 aa). A comparison of the nt and aa sequences of CPV-2b-K5-TR and CPV-2b-O1-TR is shown in Supplementary Table S2. Phylogenetic analysis based on complete genome sequences showed that the two CPV-2b isolates grouped in a cluster with Australian and East Asian isolates (Supplementary Fig. S1).
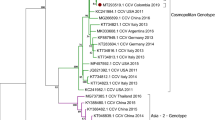
The amino acid substitutions in the VP2 protein of the isolates examined in this study are shown in Table 2. A phylogenetic tree based on VP2 nt sequences is shown in Fig. 2.
Phylogeny of complete VP2 sequences of CPV-2. The tree was constructed using the maximum-likelihood method. Bootstrap support values based on 1000 replicates are shown at the nodes. Black circles represent the CPV-2b isolates from the present study. Others are FPV and CPV-2 strains or isolates from different countries.
The amino acid substitutions in the NS1 protein are listed in Supplementary Table S3. A phylogenetic tree based on NS1 sequences showed that the two Turkish CPV-2b isolates grouped into different clusters (Supplementary Fig. S2).
Virus isolation
A mild cytopathic effect in the form of rounding, increased granularity, and detached cells could be seen 3–4 days postinfection for 37 samples at the third-passage level.
Of the 37 CPV-2 isolates obtained, two with different VP2 sequences were randomly selected, and their complete genomes were sequenced.
Statistical analysis
According to the chi-squared test analysis, breed, age, gender, and vaccination were not associated with CPV-2 infection in this study (p > 0.05) (Supplementary Table S4).
Discussion
Canine parvovirus (CPV) is an important, contagious, commonly diagnosed, and fatal pathogen of domestic and wild dogs. Age, breed, sex, stress, coinfection with another pathogen, non-vaccination, immunosuppression, and geographic region/environment have been put forward as predisposing factors for CPV-2 infection [2, 24, 41]. Other viruses, such as CCoV and CCV, which can exacerbate the clinical signs of CPV-2 infection and gastrointestinal disorders in dogs, were simultaneously investigated in this study [42]. No samples were positive for either CCoV or CCV. Several factors, such as incorrect sampling time and low viral load, could have been responsible for this finding.
CPV-2 infections are most common in puppies under the age of 6 months due to a decrease in maternal antibodies [24]. This predisposing factor is reflected in the current study with the majority of CPV-2-positive cases (42/48, 87.5%) involving 1- to 6-month-old puppies. The interference of maternal antibodies in puppies could be influenced by improper vaccination protocols, the efficiency of the immune system of puppies, and the interaction between maternal antibodies and the vaccine, causing a lack of maternal immunity [43, 44]. CPV-2b was detected in a 5-year-old adult dog with a completed CPV vaccination history, although no virus could be isolated. This negative result, which is often seen in the CPV-2b and 2c literature, could be due to the physiological decline of protective immunity or the increase in virulence and tissue tropism of specific circulating CPV strains [21, 45,46,47].
Modified live attenuated vaccines containing the original CPV-2 strain (Nobivac-Parvo C, Intervet) or CPV-2b strain (Biocan, Bioveta) are widely used in Turkey. The number of vaccinated dogs with parvoviral enteritis in this study constituted almost a quarter of the cases (15/62, 24.1%). The increase in CPV-2 cases in vaccinated dogs in Turkey or other countries may be due to vaccines containing heterologous strains or immune escape mutations of field viruses [48, 49]. Studies involving cross-protection experiments are expected to come to the fore in the future, and vaccines administered to dogs should carry dominant local variants with new epitopes in the CPV-2 capsid protein. Incorporation of field strains in commercial vaccines may be considered for effective control of canine parvovirus infection in the country.
Cases of CPV-2 occurring in vaccinated dogs might also indicate that incomplete vaccination of puppies is not sufficient to protect against CPV-2. Therefore, Turkish veterinarians should encourage pet owners to complete vaccination of puppies according to World Small Animal Veterinary Association (WSAVA) guidelines [48].
The evolution of CPV-2 has raised questions regarding the efficacy of vaccination. Similar to previous studies [7], this study identified universal mutations in the VP2 protein that distinguish canine parvoviruses from feline parvoviruses: K80R, K93N, V103A, D323N, N564S, and A568G. In addition, the aa substitutions M87L, I101T, A300G, and D305Y, which are characteristic of type 2a/2b/2c CPVs, and N426D, which is characteristic of 2b, were obtained. Other potentially important substitutions found in CPV isolates were F267Y, Y324I, and T440A, which are located in protuberances of their respective loops in the main VP2 antigenic sites [50, 51]. These mutations are associated with selective pressure and vaccine failure [7]. Strains encoding 267Y first appeared in 2002 and have become predominant since 2014 [7]. Zhou et al. [7] reported that aa residue 267, which in a 3D model of VP2 was not part of a loop structure, has the potential to cause vaccine failure. In 2006, the mutation Y324I in the VP2 protein of CPV-2, which lies adjacent to residue 323, rapidly spread amongst circulating CPV strains [7,8,9]. Evolutionary literature has associated residue 323 with changes in host range by influencing the ability of CPV to bind to the canine transferrin receptor. The mutation Y324I has also been shown to occur in regions that are associated with important antigenic epitopes [51]. The Y324I mutation in CPV-2a strains has been reported in China [52], South Korea [53], Thailand [54], Japan [55], Uruguay [56], Taiwan [57], and India [58]. The substitution Y324I was detected in all CPV-2b strains and some East Asian CPV-2 strains among the 2b/2c/new2b subtypes examined in this study. These results suggest that the Y324I substitution in CPV-2 strains is common in Asian countries [52, 59]. In the VP2 protein of CPV-2, the amino acid residue at position 440 is located in the GH loop [60]. Strains encoding 440A first appeared in 1993 and have appeared continuously in circulating CPV strains since 2005 [7]. At present, the T440A substitution is being reported frequently worldwide, and it was found in all CPV-2b strains detected in this study [7, 61, 62]. The findings of the present study clearly demonstrate that Y324I and T440A mutant CPV-2b strains have become dominant in Turkey, which is a gateway between Asia and Europe. This may contribute to virus immune escape via antigenic drift and consequent vaccine failure [7].
In addition, 5A, 13P, 52T, and 55Q mutations were detected in the VP2 protein. The 5A mutation has been detected frequently in Chinese CPV-2c isolates [61, 63], while the 13P mutation has been detected in Italian CPV-2b isolates [60]. Other substitutions seem to have emerged recently. The results show that CPV-2b continues to evolve, and further work is needed to better understand the phenotypic impact of novel aa substitutions.
The NS1 protein of CPV-2b-O1-TR was found to contain new aa substitutions, A255G and Q307T, in the origin of replication (ORI) binding domain and helicase domain, respectively. The CPV-2b-K5-TR isolate similarly showed the substitutions D309E and E375G in the helicase domain. However, a previous study by Mira et al. [64] showed different (60V, 544F, 545F, and 630P) substitutions in the NS1 protein in Asian CPV strains that were not found in this study. Limited literature is available on the NS gene of CPV-2 compared to the CPV-2 VP2 gene, and there is a lack of sequence data in GenBank for review. One recent NS1 study investigated T598 and T601 phosphorylation sites of NS1 and determined that both of these sites are crucial for viral replication and pathogenicity [65]. Further studies are needed to determine the contribution of the substitutions identified here to the biological properties of NS1.
Phylogenetic analysis revealed that the isolates CPV-2a/2b/2c/new2b and CPV-2b/c/new2b formed separate subclusters. Isolates located in the same phylogenetic cluster with different types of CPV-2 variants can be distinguished based on amino acid differences at position 426. When the VP2 phylogenetic tree was examined, some isolates were relatively closely related, while in the NS1 tree they appeared distantly related. Considering selection pressures or recombination in assessing these two differences may help to explain the conflicting phylogenetic trees obtained for VP2 and NS1 [66].
According to epidemiological surveys, the distribution of CPV-2 variants varies according to geographical region, and the dominant circulating CPV-2 variant can change over time. CPV-2b is common in Brazil, the USA, Japan, Switzerland, and South Africa, while CPV-2c is common in the United Kingdom, Vietnam, Spain, South America (except Brazil), and North America. In contrast, CPV-2a was reported to be the common antigenic type in France, Taiwan, and Italy, while both CPV-2a and CPV-2b were reported to be distributed equally in Spain [67]. Typing field isolates from different geographic regions is essential for controlling the spread of variants, understanding virus evolution, and developing strategies for control and treatment. Although subtypes 2a, 2b, and 2c have been reported in Turkey [36, 37, 40], this study shows that CPV-2b is widely distributed in Turkey as a single variant.
In conclusion, CPV-2b was found to be predominant in symptomatic dogs with CPV-2 across three different regions of Turkey. The strains included Y324I and T440A substitutions, which commonly contribute to virus immune escape. This is the first study in which complete genomic analysis of Turkish CPV-2 isolates was performed.
The evolution of CPV-2 has raised questions about the efficacy of vaccination. Therefore, continuous monitoring of the emergence and spread of new CPV-2 variants should be the primary objective of ongoing research. General control measures should be taken to reduce the prevalence of the disease in canine populations, such as adequate feeding of young dogs, control of enteric pathogens, environmental sanitation, and physical isolation of infected animals. Vaccination against CPV-2 is regarded as the primary prophylactic strategy for controlling this disease. Turkish veterinarians should inform animal owners and be encouraged to continue administering vaccines.
References
Decaro N, Buonavoglia C (2012) Canine parvovirus—a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet Microbiol 155:1–12. https://doi.org/10.1016/j.vetmic.2011.09.007
Goddard A, Leisewitz AL (2010) Canine parvovirus. Vet Clin N Am Small Anim Pract 40:1041–1053. https://doi.org/10.1016/j.cvsm.2010.07.007
Prittie J (2004) Canine parvoviral enteritis: a review of diagnosis, management, and prevention. J Vet Emerg Crit Care 13:167–176. https://doi.org/10.1111/j.1534-6935.2004.04020.x
Shackelton LA, Parrish CR, Truyen U, Holmes EC (2005) High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci USA 102:379–384. https://doi.org/10.1073/pnas.0406765102
Allison AB, Kohler DJ, Ortega A, Hoover EA, Grove DM, Holmes EC, Parrish CR (2014) Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLOS Pathog 10:e1004475. https://doi.org/10.1371/journal.ppat.1004475
Parrish CR (2011) Parvoviruses. In: Dubovi EJ, Maclachlan JN (eds) Fenner’s veterinary virology, 5th edn. Academic press, Massachusetts, pp 224–235. https://doi.org/10.1016/B978-0-12-375158-4.00007-9
Zhou P, Zeng W, Zhang X, Li S (2017) The genetic evolution of canine parvovirus—a new perspective. PLoS ONE 12:e0175035. https://doi.org/10.1371/journal.pone.0175035
Wang X, Zhang J, Huo S, Zhang Y, Wu F, Cui D, Yu H, Zhong F (2020) Development of a monoclonal antibody against canine parvovirus NS1 protein and investigation of NS1 dynamics and localization in CPV-infected cells. Protein Expr Purif 174:105682. https://doi.org/10.1016/j.pep.2020.105682
Hueffer K, Parker JS, Weichert WS, Geisel RE, Sgro JY, Parrish CR (2003) The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol 77:1718–1726. https://doi.org/10.1128/jvi.77.3.1718-1726.2003
Nelson CD, Palermo LM, Hafenstein SL, Parrish CR (2007) Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the fab fragments of monoclonal antibodies. Virology 361:283–293. https://doi.org/10.1016/j.virol.2006.11.032
Parrish CR, Aquadr CF, Strassheim M, Evermann J, Sgro J, Mohammed H (1991) Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol 65:6544–6552. https://doi.org/10.1128/JVI.65.12.6544-6552
Hoelzer K, Shackelton LA, Parrish CR, Holmes EC (2008) Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J Gen Virol 89:2280–2289. https://doi.org/10.1099/vir.0.2008/002055-0
Cotmore SF, Agbandje-McKenna M, Canuti M et al (2019) ICTV virus taxonomy profile: parvoviridae. J Gen Virol 100:367–368. https://doi.org/10.1099/jgv.0.001212
Stucker KM, Pagan I, Cifuente JO, Kaelber JT, Lillie TD, Hafenstein S, Holmes EC, Parrish CR (2012) The role of evolutionary intermediates in the host adaptation of canine parvovirus. J Virol 86:1514–1521. https://doi.org/10.1128/jvi.06222-11
Hoelzer K, Parrish CR (2010) The emergence of parvoviruses of carnivores. Vet Res 41:39. https://doi.org/10.1051/vetres/2010011
Decaro N, Desario C, Addie DD et al (2007) The study molecular epidemiology of canine parvovirus, Europe. Emerg Infect Dis 13:1222–1224
Zhao H, Wang J, Jiang Y, Cheng Y, Lin P, Zhu H, Han G, Yi L, Zhang S, Guo L, Cheng S (2015) Typing of canine parvovirus strains circulating in North-East China. Transbound Emerg Dis 64:495–503. https://doi.org/10.1111/tbed.12390
Nandi S, Chidri S, Kumar M, Chauhan RS (2010) Occurrence of canine parvovirus type 2c in the dogs with haemorrhagic enteritis in India. Res Vet Sci 88:169–171
Perez R, Francia L, Romero V, Maya L, Lopez I, Hernandez M (2007) First detection of canine parvovirus type 2c in South America. Vet Microbiol 124:147–152
Decaro N, Desario C, Campolo M, Elia G, Martella V, Ricci D, Lorusso E, Buonavoglia C (2005) Clinical and virological findings in pups naturally infected by canine parvovirus type 2 Glu-426 mutant. J Vet Diagn Invest 17:133–138. https://doi.org/10.1177/104063870501700206
Decaro N, Desario C, Elia G, Martella V, Mari V, Lavazza A, Nardi M, Buonavoglia C (2008) Evidence for immunisation failure in vaccinated adult dogs infected with canine parvovirus type 2c. New Microbiol 31:125–130
Ohshima T, Hisaka M, Kawakami K, Kishi M, Tohya Y, Mochizuki M (2008) Chronological analysis of canine parvovirus type 2 isolates in Japan. J Vet Med Sci 70:769–775. https://doi.org/10.1292/jvms.70.769
Martella V, Decaro N, Elia G, Buonavoglia C (2005) Surveillance activity for canine parvovirus in Italy. J Vet Med B Infect Dis Vet Public Health 52:312–315. https://doi.org/10.1111/j.1439-0450.2005.00875.x
Mylonakis ME, Kalli I, Rallis TS (2016) Canine parvoviral enteritis: an update on the clinical diagnosis, treatment, and prevention. Vet Med 7:91–100. https://doi.org/10.2147/VMRR.S80971
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (2012) Virus taxonomy: classification and nomenclature of viruses. In: Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA
Greene CE, Decaro N (2012) Canine viral enteritis. In: Greene CE (ed) Infectious diseases of the dog and cat, 4th edn. Elsevier Saunders, St. Louis, pp 67–80
Todd D, Mcnulty MS, Adair BM, Allan GM (2001) Animal circoviruses. Adv Virus Res 57:1–70. https://doi.org/10.1016/S0065-3527(01)57000-1
Li L, McGraw S, Zhu K, Leutenegger CM, Marks SL, Kubiski S, Gaffney P, Dela Cruz FN Jr, Wang C, Delwart E, Pesavento PA (2013) Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis 19:534–541. https://doi.org/10.3201/eid1904.121390
Decaro N, Martella V, Desario C, Lanave G, Circella E, Cavalli A, Elia G, Camero M, Buonavoglia C (2014) Genomic characterization of a circovirus associated with fatal hemorrhagic enteritis in dog, Italy. PLoS ONE 9:e105909. https://doi.org/10.1371/journal.pone.0105909
Vannamahaxay S, Vongkhamchanh S, Intanon M, Tangtrongsup S, Tiwananthagorn S, Pringproa K, Chuammitri P (2017) Molecular characterization of canine parvovirus in Vientiane, Laos. Arch Virol 162:1355–1361. https://doi.org/10.1007/s00705-016-3212-1
Canuti M, Britton AP, Graham SM, Lang AS (2017) Epidemiology and molecular characterization of protoparvoviruses infecting wild raccoons (Procyon lotor) in British Columbia, Canada. Virus Res 242:85–89. https://doi.org/10.1016/j.virusres.2017.09.015
Pratelli A, Decaro N, Tinelli A, Martella V, Elia G, Tempesta M, Cirone F, Buonavoglia C (2004) Two genotypes of canine coronavirus simultaneously detected in the fecal samples of dogs with diarrhea. J Clin Microbiol 42:1797–1799. https://doi.org/10.1128/JCM.42.4.1797-1799.2004
Kotsias F, Bucafusco D, Nuñez DA, Lago Borisovsky LA, Rodriguez M, Bratanich AC (2019) Genomic characterization of canine circovirus associated with fatal disease in dogs in South America. PLoS ONE 14:e0218735. https://doi.org/10.1371/journal.pone.0218735
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Isidan H, Turan T (2021) A comprehensive study of canine parvoviruses (Carnivore protoparvovirus 1, Carnivore bocaparvovirus 1 and 2) from shelter dogs in Turkey. Vet Med-Czech 66:423–430. https://doi.org/10.17221/130/2020-VETMED
Timurkan M, Oğuzoğlu T (2015) Molecular characterization of canine parvovirus (CPV) infection in dogs in Turkey. Vet Ital 51:39–44. https://doi.org/10.12834/VetIt.263.908.3
Polat PF, Şahan A, Aksoy G, Timurkan MO, Dinçer E (2019) Molecular and restriction fragment length polymorphism analysis of canine parvovirus 2 (CPV-2) in dogs in southeast Anatolia, Turkey. Onderstepoort J Vet Res 86:e1–e8. https://doi.org/10.4102/ojvr.v86i1.1734
Dincer E (2017) Molecular characterization and phylogenetic analysis of canine parvovirus 2 in dogs, Mersin Province, Turkey. Etlik Veteriner Mikrobiyoloji Dergisi 28:96–100
Yilmaz Z, Pratelli A, Torun S (2005) Distribution of antigen types of canine parvovirus type 2 in dogs with hemorrhagic enteritis in Turkey. Turk J Vet Anim Sci 29:1073–1076
Kalli I, Leontides LS, Mylonakis ME, Adamama-Moraitou K, Rallis T, Koutinas AF (2010) Factors affecting the occurrence, duration of hospitalization and final outcome in canine parvovirus infection. Res Vet Sci 89:174–178. https://doi.org/10.1016/j.rvsc.2010.02.013
Pratelli A, Tempesta M, Roperto FP, Sagazio P, Carmichael L, Buonavoglia C (1999) Fatal coronavirus infection in puppies following canine parvovirus 2b infection. J Vet Diagn Invest 11:550–553. https://doi.org/10.1177/104063879901100615
Klingborg DJ, Hustead DR, Curry-Galvin EA et al (2002) AVMA Council on Biologic and Therapeutic Agents’ report on cat and dog vaccines. J Am Vet Med Assoc 221:1401–1407. https://doi.org/10.2460/javma.2002.221.1401
Umar S, Ali A, Younus M, Maan MK, Ali S, Khan A, Irfan M (2015) Prevalence of canine parvovirus infection at different pet clinics in Lahore, Pakistan. Pakistan J Zool 47:657–663
Calderon MG, Mattion N, Bucafusco D, Fogel F, Remorini P, La Torre J (2009) Molecular characterization of canine parvovirus strains in Argentina: Detection of the pathogenic variant CPV2c in vaccinated dogs. J Virol Methods 159:141–145. https://doi.org/10.1016/j.jviromet.2009.03.013
Cavalli A, Bozzo G, Decaro N, Tinelli A, Aliberti A, Buonavoglia D (2001) Characterization of a canine parvovirus strain isolated from an adult dog. Microbiologica 24:239–242
Cavalli A, Martella V, Desario C, Camero M, Bellacicco AL, De Palo P, Decaro N, Elia G, Buonavoglia C (2008) Evaluation of the antigenic relationships among canine parvovirus type 2 variants. Clin Vaccine Immunol 15:534–539. https://doi.org/10.1128/CVI.00444-07
Day MJ, Horzinek MC, Schultz RD, Squires RA (2016) Vaccination Guidelines Group (VGG) of the World Small Animal Veterinary Association (WSAVA). WSAVA Guidelines for the vaccination of dogs and cats. J Small Anim Pract 57:E1–E45. https://doi.org/10.1111/jsap.2_12431
Mittal M, Chakravarti S, Mohapatra JK, Chug PK, Dubey R, Upmanuyu V, Narwal PS, Kumar A, Churamani CP, Kanwar NS (2014) Molecular typing of canine parvovirus strains circulating from 2008 to 2012 in an organized kennel in India reveals the possibility of vaccination failure. Infect Genet Evol 23:1–6. https://doi.org/10.1016/j.meegid.2014.01.015
Tsao J, Chapman MS, Agbandje M et al (1991) The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456–1464. https://doi.org/10.1126/science.2006420
Hueffer K, Parrish CR (2003) Parvovirus host range, cell tropism and evolution. Curr Opin Microbiol 6:392–398. https://doi.org/10.1016/s1369-5274(03)00083-3
Zhang R, Yang S, Zhang W, Zhang T, Xie Z, Feng H, Wang S, Xia X (2010) Phylogenetic analysis of the VP2 gene of canine parvoviruses circulating in China. Virus Genes 40:397–402. https://doi.org/10.1007/s11262-010-0466-7
Yoon SH, Jeong W, Kim HJ, An DJ (2009) Molecular insights into the phylogeny of canine parvovirus 2 (CPV-2) with emphasis on Korean isolates: a Bayesian approach. Arch Virol 154:1353–1360. https://doi.org/10.1007/s00705-009-0444-3
Phromnoi S, Sirinarumitr K, Sirinarumitr T (2010) Sequence analysis of VP2 gene of canine parvovirus isolates in Thailand. Virus Genes 41:23–29. https://doi.org/10.1007/s11262-010-0475-6
Soma T, Taharaguchi S, Ohinata T, Ishii H, Hara M (2013) Analysis of the VP2 protein gene of canine parvovirus strains from affected dogs in Japan. Res Vet Sci 94:368–371. https://doi.org/10.1016/j.rvsc.2012.09.013
Pérez R, Bianchi P, Calleros L, Francia L, Hernández M, Maya L, Panzera Y, Sosa K, Zoller S (2012) Recent spreading of a divergent canine parvovirus type 2a (CPV-2a) strain in a CPV-2c homogenous population. Vet Microbiol 155:214–219. https://doi.org/10.1016/j.vetmic.2011.09.017
Chou SJ, Lin HT, Wu JT, Yang WC, Chan KW (2013) Genotyping of canine parvovirus type 2 VP2 gene in southern Taiwan in 2011. Taiwan Vet J 39:81–92
Mukhopadhyay HK, Matta SL, Amsaveni S, Antony PX, Thanislass J, Pillai RM (2014) Phylogenetic analysis of canine parvovirus partial VP2 gene in India. Virus Genes 201448:89–95. https://doi.org/10.1007/s11262-013-1000-5
Lin CN, Chien CH, Chiou MT, Chueh LL, Hung MY, Hsu HS (2014) Genetic characterization of type 2a canine parvoviruses from Taiwan reveals the emergence of an Ile324 mutation in VP2. Virol J 11:39. https://doi.org/10.1186/1743-422X-11-39
Battilani M, Modugno F, Mira F, Purpari G, Di Bella S, Guercio A, Balboni A (2019) Molecular epidemiology of canine parvovirus type 2 in Italy from 1994 to 2017: recurrence of the CPV-2b variant. BMC Vet Res 15:393. https://doi.org/10.1186/s12917-019-2096-1
Wang J, Lin P, Zhao H, Cheng Y, Jiang Z, Zhu H, Wu H, Cheng S (2016) Continuing evolution of canine parvovirus in China: isolation of novel variants with an Ala5Gly mutation in the VP2 protein. Infect Genet Evol 38:73–78. https://doi.org/10.1016/j.meegid.2015.12.009
Hong C, Decaro N, Desario C, Tanner P, Pardo MC, Sanchez S, Buonavoglia C, Saliki JT (2007) Occurrence of canine parvovirus type 2c in the United States. J Vet Diagn Invest 19(5):535–539. https://doi.org/10.1177/104063870701900512
Zhuang QY, Qiu Y, Pan ZH, Wang SC, Wang B, Wu WK, Yu JM, Yi Y, Sun FL, Wang KC (2019) Genome sequence characterization of canine parvoviruses prevalent in the Sichuan province of China. Transbound Emerg Dis 66:897–907. https://doi.org/10.1111/tbed.13100
Mira F, Canuti M, Purpari G et al (2019) Molecular characterization and evolutionary analyses of carnivore protoparvovirus 1 NS1 gene. Viruses 11:308. https://doi.org/10.3390/v11040308
Miao B, Chen S, Zhang X et al (2022) T598 and T601 phosphorylation sites of canine parvovirus NS1 are crucial for viral replication and pathogenicity. Vet Microbiol 264:109301. https://doi.org/10.1016/j.vetmic.2021.109301
Wang H, Jin H, Li Q et al (2016) Isolation and sequence analysis of the complete NS1 and VP2 genes of canine parvovirus from domestic dogs in 2013 and 2014 in China. Arch Virol 161:385–393. https://doi.org/10.1007/s00705-015-2620-y
Nandi S, Kumar M (2010) Canine parvovirus: current perspective. Indian J Virol 21:31–44. https://doi.org/10.1007/s13337-010-0007-y
Funding
This study did not receive funding from any institution or organization.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by HA, OA, and KÇT. Analysis was performed by HA, ST, KC-S. The first draft of the manuscript was written by HA, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
No attempt was made to adversely affect animal health or disrupt the tissue integrity of the dogs selected in this study. All procedures performed in animal studies were in compliance with the institution (Fırat University Animal Experiments Local Ethics Committee) and international ethical standards.
Informed consent
The consent of animal owners was obtained at the time of sampling.
Additional information
Handling Editor: Ana Cristina Bratanich .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abayli, H., Aslan, O., Tumer, K.C. et al. Predominance and first complete genomic characterization of canine parvovirus 2b in Turkey. Arch Virol 167, 1831–1840 (2022). https://doi.org/10.1007/s00705-022-05509-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05509-4