Abstract
Main conclusion
SbCASP4 improves the salt tolerance of sweet sorghum [Sorghum bicolor (L.) Mocnch] by enhancing the root apoplastic barrier and blocking the transport of sodium ions to the shoot.
Abstract
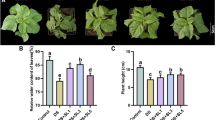
Sweet sorghum [Sorghum bicolor (L.) Mocnch] is a C4 crop with high biomass and tolerance to abiotic stresses such as salt, drought, and waterlogging. Sweet sorghum is widely used in bioenergy production, as a forage crop, and in liquors and beer. Root salt exclusion has been reported to underlie the salt tolerance of sweet sorghum. The Casparian strip has a key role in root salt exclusion, and the membrane domain protein (CASP) family participates in Casparian strip aggregation. However, the function and the regulatory mechanisms of SbCASP in response to salt stress in sweet sorghum are unclear. In the current study, we cloned SbCASP4 and determined that it is induced by salt stress and expressed in the endodermis cells of sweet sorghum. Histochemical staining and physiological indicators showed that heterologous expression of SbCASP4 significantly increased the tolerance to salt stress in transgenic Arabidopsis thaliana. Compared with wild type and casp5 mutants, under 50 mM NaCl treatment, SbCASP4-expression lines had the less leaf Na+, lower PI accumulation in stele, smaller oxidative damage and higher salinity threshold, longer root length and higher expression levels of the genes related to Casparian strip formation.







Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Andersen TG, Barberon M, Geldner N (2015) Suberization-the second life of an endodermal cell. Curr Opin Plant Biol 28:9–15. https://doi.org/10.1016/j.pbi.2015.08.004
Baxter I, Hosmani PS, Rus A, Lahner B, Borevitz JO, Muthukumar B, Mickelbart MV, Schreiber L, Franke RB, Salt DE (2009) Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet 5(5):e1000492. https://doi.org/10.1371/journal.pgen.1000492
Blumwald E (2000) Sodium transport and salt tolerance in plants. Cur Opin Cell Biol 12(4):431–434. https://doi.org/10.1016/S0955-0674(00)00112-5
Cheeseman JM (1982) Pump-leak sodium fluxes in low salt corn roots. J Membr Biol 70(2):157–164. https://doi.org/10.1007/BF01870225
Chen M, Yang Z, Liu J, Zhu TT, Wei XC, Fan H, Wang BS (2018) Adaptation mechanism of salt excluders under saline conditions and its applications. Int J Mol Sci 19(11):13. https://doi.org/10.3390/ijms19113668
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743. https://doi.org/10.1002/ece3.6625
Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F (2009) CYP86B1 is required for very long chain omega-hydroxyacid and alpha, omega -dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol 150(4):1831–1843. https://doi.org/10.1104/pp.109.141408
Daniele R, Bert DR, Valérie Dénervaud T, Alexandre P, Julien A, Vermeer JEM, Misako Y, York-Dieter S, Tom B, Niko G (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473(7347):380–383. https://doi.org/10.1038/nature10070
Ding TL, Yang Z, Wei XC, Yuan F, Yin SS, Wang BS (2018) Evaluation of salt-tolerant germplasm and screening of the salt-tolerance traits of sweet sorghum in the germination stage. Funct Plant Biol 45(10):1073–1081. https://doi.org/10.1071/fp18009
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7(7):1085. https://doi.org/10.1105/tpc.7.7.1085
Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE (2013) Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. P Natl Acad Sci 110(35):14498. https://doi.org/10.1073/pnas.1308412110
Hui Z, Tian F-X, Wang G-k, Wang G-P, Wang W (2012) The antioxidative defense system is involved in the delayed senescence in a wheat mutant tasg1. Plant Cell Rep 31(6):1073–1084. https://doi.org/10.1007/s00299-012-1226-z
Ines CRB, Nelson R-M, Niko G (2019) The Casparian strip—one ring to bring cell biology to lignification? Cur Opin Biotech 56:121–129. https://doi.org/10.4161/psb.6.10.17054
Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, Tanoi K, Matsumura H, Fujii-Kashino M, Horiuchi T, Nayef MA, Shabala S, An G, Ma JF, Horie T (2017) OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J 91(4):657–670. https://doi.org/10.1111/tpj.13595
Krishnamurthy P, Ranathunge K, Franke R, Prakash HS, Schreiber L, Mathew MK (2009) The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta 230(1):119–134. https://doi.org/10.1007/s00425-009-0930-6
Krishnamurthy P, Ranathunge K, Nayak S, Schreiber L, Mathew MK (2011) Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.). J Exp Bot 62(12):4215–4228. https://doi.org/10.1093/jxb/err135
Krishnamurthy P, Jyothi-Prakash P, Qin L, He J, Lin Q, Loh C-S, Kumar P (2014) Role of root hydrophobic barriers in salt exclusion of a mangrove plant Avicennia officinalis. Plant Cell Environ. https://doi.org/10.1111/pce.12272
Lee Y, Rubio MC, Alassimone J, Geldner N (2013) A mechanism for localized lignin deposition in the endodermis. Cell 153(2):402–412. https://doi.org/10.1016/j.cell.2013.02.045
Leng B, Wang X, Yuan F, Zhang H, Lu C, Chen M, Wang B (2021) Heterologous expression of the Limonium bicolor MYB transcription factor LbTRY in Arabidopsis thaliana increases salt sensitivity by modifying root hair development and osmotic homeostasis. Plant Sci 302:110704. https://doi.org/10.1016/j.plantsci.2020.110704
Li P, Yu Q, Gu X, Xu C, Qi S, Wang H, Zhong F, Baskin TI, Rahman A, Wu S (2018) Construction of a functional casparian strip in non-endodermal lineages is orchestrated by two parallel signaling systems in Arabidopsis thaliana. Curr Biol 28(17):2777-2786.e2772. https://doi.org/10.1016/j.cub.2018.07.028
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59(1):651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. P Natl Acad Sci USA 109(25):10101–10106. https://doi.org/10.1073/pnas.1205726109
Ochiai K, Matoh T (2002a) Characterization of the Na+ delivery from roots to shoots in rice under saline stress: excessive salt enhances apoplastic transport in rice plants. Soil Sci Plant Nutr 48(3):371–378. https://doi.org/10.1080/00380768.2002.10409214
Ochiai K, Matoh T (2002b) Characterization of the Na+ delivery from roots to shoots in rice under saline stress: excessive salt enhances apoplastic transport in rice plants. Soil Sci Plant Nutr 48:371–378. https://doi.org/10.1080/00380768.2002.10409214
Robinson SJ, Tang LH, Mooney BA, McKay SJ, Clarke WE, Links MG, Karcz S, Regan S, Wu Y-Y, Gruber MY (2009) An archived activation tagged population of Arabidopsis thaliana to facilitate forward genetics approaches. BMC Plant Biol 9(1):101. https://doi.org/10.1186/1471-2229-9-101
Roppolo D, De RB, Dénervaud TV, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473(7347):380–383. https://doi.org/10.1038/nature10070
Rossi L, Francini A, Minnocci A, Sebastiani L (2015) Salt stress modifies apoplastic barriers in olive (Olea europaea L.): a comparison between a salt-tolerant and a salt-sensitive cultivar. Sci Hortic 192:38–46. https://doi.org/10.1016/j.scienta.2015.05.023
Sakuraba Y, Bülbül S, Piao W, Choi G, Paek N-C (2017) Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J 92(6):1106–1120. https://doi.org/10.1111/tpj.13747
Schreiber L, Hartmann K, Skrabs M, Zeier J (1999) Apoplastic barriers in roots: chemical composition of endodermal and hypodermal cell walls. J Exp Bot 50(337):1267–1280. https://doi.org/10.1093/jexbot/50.337.1267
Sewalt VJ, Ni W, Blount JW, Jung HG, Masoud SA, Howles PA, Lamb C, Dixon RA (1997) Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of L-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol 115:41–50. https://doi.org/10.1104/pp.115.1.29
Shi H, Ishitani M, Kim C, Zhu J-K (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci 97(12):6896–6901. https://doi.org/10.1073/pnas.120170197
Sui N, Yang Z, Liu M, Wang B (2015) Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16(1):534. https://doi.org/10.1186/s12864-015-1760-5
Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi NI, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, Schroeder JI, Ma JF, Horie T (2016) OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol 16(1):22. https://doi.org/10.1186/s12870-016-0709-4
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. J Plant Physiol 153(3):895–905. https://doi.org/10.1104/pp.110.155119
Wang Z, Yamaji N, Huang S, Zhang X, Shi M, Fu S, Yang G, Feng J, Xia J (2019) OsCASP1 is required for Casparian strip formation at endodermal cells of rice roots for selective uptake of mineral elements. Plant Cell 31(11):2636–2648. https://doi.org/10.1105/tpc.19.00296
Wang Z, Shi M, Wei Q, Chen Z, Huang J, Xia J (2020) OsCASP1 forms complexes with itself and OsCASP2 in rice. Plant Signal Behav. https://doi.org/10.1080/15592324.2019.1706025
Wei X, Yan X, Yang Z, Han G, Wang L, Yuan F, Wang B (2020a) Salt glands of recretohalophyte Tamarix under salinity: their evolution and adaptation. Ecol Evol 10(17):9384–9395. https://doi.org/10.1002/ece3.6625
Wei X, Yang Z, Han G, Zhao X, Yin S, Yuan F, Wang B (2020b) The developmental dynamics of the sweet sorghum root transcriptome elucidate the differentiation of apoplastic barriers. Plant Signal Behav. https://doi.org/10.1080/15592324.2020.1724465
Yang Z, Zheng HX, Wei XC, Song J, Wang BS, Sui N (2018) Transcriptome analysis of sweet Sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil 430(1–2):423–439. https://doi.org/10.1007/s11104-018-3736-0
Yeo A, Yeo M, Flowers T (1987) The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J Exp Bot 38(7):1141–1153. https://doi.org/10.1093/jxb/38.7.1141
Yuan F, Leng BY, Wang BS (2016) Progress in studying salt secretion from the salt glands in recretohalophytes: how do plants secrete salt? Front Plant Sci 7(977):977. https://doi.org/10.3389/fpls.2016.00977
Yuan F, Xu Y, Leng B, Wang B (2019) Beneficial effects of salt on halophyte growth: morphology, cells, and genes. Open Life Sci 14(1):191–200
Zeier J, Goll A, Yokoyama M, Karahara I, Schreiber L (1999) Structure and chemical composition of endodermal and rhizodermal hypodermal walls of several species. Plant Cell Envir 22(3):271–279. https://doi.org/10.1046/j.1365-3040.1999.00401.x
Zhao C, Zhang H, Song C, Zhu J, Shabala S (2020) Mechanisms of plant responses and adaptation to soil salinity. Innov 1(1):100017. https://doi.org/10.1016/j.xinn.2020.100017
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6(2):66–71. https://doi.org/10.1016/S1360-1385(00)01838-0
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6(5):441–445. https://doi.org/10.1016/S1369-5266(03)00085-2
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167(2):313–324. https://doi.org/10.1016/j.cell.2016.08.029
Acknowledgements
This work was supported by the NSFC (National Natural Science Research Foundation of China; project nos. 31770288 and 31600200), the Shandong Province Key Research and Development Plan (2017CXGC0313), and the Natural Science Research Foundation of Shandong Province (ZR2014CZ002; ZR2017MC003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

Supplementary file1 Figure S1. The map of vectors: pCAMBIA 1300 (a); pCAMBIA 3301 (b). (JPG 1341 KB)

425_2021_3731_MOESM2_ESM.jpg
Supplementary file2 Figure S2. Temporal expression pattern of SbCASP4 in sweet sorghum roots (M-81E) at stage I and II under150 mM NaCl. (JPG 2000 KB)

425_2021_3731_MOESM3_ESM.jpg
Supplementary file3 Figure S3. Phylogenetic tree of sorghum and Arabidopsis CASP genes (a), and phylogenetic tree of sorghum and rice CASP genes (b). (JPG 4173 KB)

425_2021_3731_MOESM4_ESM.jpg
Supplementary file4 Figure S4. Identification of positive Arabidopsis transgenic lines. a: PCR identification of 35S:SbCASP4-GFP-positive lines. b: The relative expression levels of SbCASP4 in different lines. Values are means ± SD (n = 3). Different letters indicate significant differences at P = 0.05 as determined by Duncan′s multiple range test. c: PCR identification of pSbCASP4:SbCASP4-GFP-positive lines. d: PCR identification of pSbCASP4:GUS-positive lines. (JPG 2795 KB)

425_2021_3731_MOESM5_ESM.jpg
Supplementary file5 Figure S5. Screening of the Arabidopsis homozygous casp5 mutants, (CS1015546) the Arabidopsis homolog of SbCASP4, and identification of T-DNA insertion sites. a: PCR identification of CS1015546 in the casp5 mutants, M: 2,000 bp DNA Marker; b: Sequencing identification of primer RP + pSKTAIL-L1; The red arrow indicates the insertion position of the T-DNA; c: Diagram of the T-DNA insertion site. (JPG 4203 KB)

425_2021_3731_MOESM6_ESM.jpg
Supplementary file6 Figure S6. K+ content of shoot (a) and root (b) of the Arabidopsis wild-type, casp5 mutant, and SbCASP4-expression lines under 0 and 50 mM NaCl for 5 days. Values are means ± SD (n = 3). Different letters indicate significant differences at P = 0.05 as determined by Duncan′s multiple range test. (JPG 2915 KB)

425_2021_3731_MOESM7_ESM.jpg
Supplementary file7 Figure S7. The relative expression levels of ion transporter genes in the Arabidopsis wild-type, casp5 mutant, and SbCASP4-expression lines under 0 and 50 mM NaCl for 5 days. SbCASP4 does not affect the expression of ion transporter genes. (a) AtSOS1; (b) AtHKT1; (c) AtNHX1. Values are means ± SD (n = 3). Different letters indicate significant differences at P = 0.05 as determined by Duncan’s multiple range test. (JPG 2400 KB)
Rights and permissions
About this article
Cite this article
Wei, X., Liu, L., Lu, C. et al. SbCASP4 improves salt exclusion by enhancing the root apoplastic barrier. Planta 254, 81 (2021). https://doi.org/10.1007/s00425-021-03731-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03731-z