Abstract
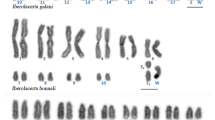
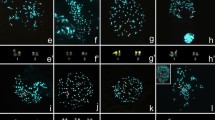
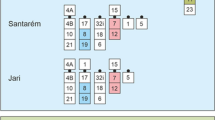
The evolutionary clade comprising Nanger, Eudorcas, Gazella, and Antilope, defined by an X;BTA5 translocation, is noteworthy for the many autosomal Robertsonian fusions that have driven the chromosome number variation from 2n = 30 observed in Antilope cervicapra, to the 2n = 58 in present Eudorcas thomsoni and Eudorcas rufifrons. This work reports the phylogenetic relationships within the Antilopini using comprehensive cytogenetic data from A. cervicapra, Gazella leptoceros, Nanger dama ruficollis, and E. thomsoni together with corrected karyotypic data from an additional nine species previously reported in the literature. Fluorescence in situ hybridization using BAC and microdissected cattle painting probes, in conjunction with differential staining techniques, provide the following: (i) a detailed analysis of the E. thomsoni chromosomes, (ii) the identification and fine-scale analysis the BTA3 orthologue in species of Antilopini, and (iii) the location of the pseudoautosomal regions on sex chromosomes of the four species. Our phylogenetic analysis of the chromosomal data supports monophyly of Nanger and Eudorcas and suggests an affiliation between A. cervicapra and some of the Gazella species. This renders Gazella paraphyletic and emphasizes a closer relationship between Antilope and Gazella than what has previously been considered.







Similar content being viewed by others
References
Ashley T (2002) X-Autosome translocations, meiotic synapsis, chromosome evolution and speciation. Cytogenet Genome Res 96:33–39
Avise JC, Robinson TJ (2008) Hemiplasy: a new term in the lexicon of phylogenetics. Syst Biol 57:503–507
Bärmann EV, Rössner GE, Wörheide G (2013) A revised phylogeny of Antilopini (Bovidae, Artiodactyla) using combined mitochondrial and nuclear genes. Mol Phylogen Evol 67:484–493
Bibi F (2013) A multi-calibrated mitochondrial phylogeny of extant Bovidae (Artiodactyla, Ruminantia) and the importance of the fossil record to systematics. BMC Evol Biol 13:166
Biémont C, Vieira C (2006) Junk DNA as an evolutionary force. Nature 443(7111):521–524
Brown JD, O’Neill RJ (2010) Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet 11:291–316
Buckland RA, Evans HJ (1978) Cytogenetic aspects of phylogeny in the Bovidae. I. G-banding. Cytogenet. Cell Genet 78:42–63
Cernohorska H, Kubickova S, Vahala J, Rubes J (2012) Molecular insights into X; BTA5 chromosome rearrangements in the tribe Antilopini (Bovidae). Cytogenet Genome Res 136:188–198
Cernohorska H, Kubickova S, Kopecna O, Kulemzina AI, Perelman PL, Elder FF, Robinson TJ, Graphodatsky AS, Rubes J (2013) Molecular cytogenetic insights to the phylogenetic affinities of the giraffe (Giraffa camelopardalis) and pronghorn (Antilocapra americana). Chromosome Res 21:447–460
Chaves R, Adega F, Heslop-Harrison JS, Guedes-Pinto H, Wienberg J (2003) Complex satellite DNA reshuffling in the polymorphic t(1;29) Robertsonian translocation and evolutionarily derived chromosomes in cattle. Cromosome Res 11:641–648
Decker JE, Pires JC, Conant GC et al (2009) Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci U S A 106:18644–18649
Dobigny G, Ozouf-Costaz C, Bonillo C, Volobouev V (2004) Viability of X-autosome translocations in mammals: an epigenomic hypothesis from a rodent case-study. Chromosoma 113:34–41
Elder FFB; Hsu TC (1988) Tandem fusion in the evolution of mammalian chromosomes. The Cytogenetics of Mammalian Autosomal Rearrangements, pages 481–501, Alan R. Liss, Inc.
Fernández MH, Vrba ES (2005) A complete estimate of the phylogenetic relationships in Ruminantia: a dated species-level supertree of the extant ruminants. Biol Rev 80:269–302
Gallagher DS Jr, Womack JE (1992) Chromosome conservation in the Bovidae. J Hered 83:287–298
Gallagher DS Jr, Davis SK, De Donato M, Burzlaff JD, Womack JE, Taylor JF, Kumamoto AT (1999) A molecular cytogenetic analysis of the tribe Bovini (Artiodactyla: Bovidae: Bovinae) with an emphasis on sex chromosome morphology and NOR distribution. Chromosome Res 7:481–492
Gatesy J, Yelon D, DeSalle R, Vrba ES (1992) Phylogeny of the Bovidae (Artiodactyla, Mammalia), based on mitochondrial ribosomal DNA sequences. J Mol Biol Evol 9:433–446
Goodpasture C, Bloom SE (1975) Visualization of nucleolar organizer in mammalian chromosomes using silver staining. Chromosoma 53:37–50
Groves C, Grubb P (2011) Ungulate taxonomy. The Johns Hopkins University Press, Baltimore
Hassanin A, Douzery EJP (1999) The tribal radiation of the family Bovidae (Artiodactyla) and the evolution of the mitochondrial cytochrome b gene. Mol Phylogenet Evol 13:227–243
Hassanin A, Delsuc F, Ropiquet A et al (2012) Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol 335:32–50
Iannuzzi L, Di Berardino D, Gustavsson I, Ferrara L, Di Meo GP (1987) Centromeric loss in translocations of centric fusion type in cattle and water buffalo. Hereditas 106:73–81
ISCNDB 2000 (2001) International system for chromosome nomenclature of domestic bovids. Cytogenet Cell Genet 92:283–299
Kejnovsky E, Hobza R, Cermak T, Kubat Z, Vyskot B (2009) The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity 102:533–541
Kidwell MG, Lisch DR (2001) Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1–24
King M (1993) Species evolution: the role of chromosome change. Cambridge University Press, Cambridge
Kopecna O, Kubickova S, Cernohorska H, Cabelova K, Vahala J, Martinkova N, Rubes J (2014) Tribe-specific satellite DNA in non-domestic Bovidae. Chromosome Res 22:277–291
Kubickova S, Cernohorska H, Musilova P, Rubes J (2002) The use of laser microdissection for the preparation of chromosome-specific painting probes in farm animals. Chromosome Res 10:571–577
Kumamoto AT, Kingswood OA, Rebholz WER, Houck ML (1995) The chromosomes of Gazella bennetti and Gazella saudiya. Int J Mammal Biol 60:159–169
Louzada S, Paço A, Kubickova S, Adega F, Guedes-Pinto H, Rubes J, Chaves R (2008) Different evolutionary trails in the related genomes Cricetus cricetus and Peromyscus eremicus (Rodentia, Cricetidae) uncovered by orthologous satellite DNA repositioning. Micron 39:1149–1155
Marcot JD (2007) Molecular phylogeny of terrestrial artiodactyls. In: Prothero DR, Foss SE (eds) The evolution of Artiodactyls. The John Hopkins University Press, Baltimore, pp 4–18
Marshall OJ, Chueh AC, Wong LH, Choo KH (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82:261–282
Matthee CA, Davis SK (2001) Molecular insights into the evolution of the family Bovidae: a nuclear DNA perspective. Mol Biol Evol 18:1220–1230
Nguyen TT, Aniskin VM, Gerbault-Seureau M, Planton H, Renard JP, Nguyen BX, Hassanin A, Volobouev VT (2008) Phylogenetic position of the saola (Pseudoryx nghetinhensis) inferred from cytogenetic analysis of 11 species of Bovidae. Cytogenet Genome Res 122:41–54
O´Brien SJ, Menninger JC, Nash WG (2006) Atlas of mammalian chromosomes. Wiley, Hoboken
Rebholz W, Harley E (1999) Phylogenetic relationships in the bovid subfamily Antilopinae based on mitochondrial DNA sequences. Mol Phylogenet Evol 12:87–94
Robinson TJ, Ruiz-Herrera A, Avise JC (2008) Hemiplasy and homoplasy in the karyotypic phylogenies of mammals. Proc Natl Acad Sci U S A 105:14477–81
Robinson TJ, Ropiquet A (2011) Examination of hemiplasy, homoplasy and phylogenetic discordance in chromosomal evolution of the Bovidae. Syst Biol 60:439–450
Robinson TJ, Cernohorska H, Diedericks G, Cabelova K, Duran A, Matthee CA (2014) Phylogeny and vicariant speciation of the Grey Rhebok, Pelea capreolus. Heredity 112:325–332
Rocchi M, Archidiacono N, Schempp W, Capozzi O, Stanyon R (2012) Centromere repositioning in mammals. Heredity 108:59–67
Rokas A, Holland PW (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15:454–459
Ropiquet A, Gerbault-Seureau M, Deuve JL, Gilbert C, Pagacova E, Chai N, Rubes J, Hassanin A (2008) Chromosome evolution in the subtribe Bovina (Mammalia, Bovidae): the karyotype of the Cambodian banteng (Bos javanicus birmanicus) suggests that Robertsonian translocations are related to interspecific hybridization. Chromosome Res 16:1107–1118
Ropiquet A, Li B, Hassanin A (2009) SuperTRI: a new approach based on branch support analyses of multiple independent data sets for assessing reliability of phylogenetic inferences. C R Biol 332:832–847
Rubes J, Kubickova S, Pagacova E et al (2008) Phylogenomic study of spiral-horned antelope by cross-species chromosome painting. Chromosome Res 16:935–947
Rubes J, Musilova P, Kopecna O, Kubickova S, Cernohorska H, Kulemsina AI (2012) Comparative molecular cytogenetics in Cetartiodactyla. Cytogenet Genome Res 137:194–207
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–25
Seabright M (1971) A rapid banding technique for human chromosomes. Lancet 2:971–972
Stimpson KM, Sullivan BA (2010) Epigenomics of centromere assembly and function. Curr Opin Cell Biol 22:772–80
Swofford DL (2002) PAUP: phylogenetic analysis using parsimony. Version 4.0b10. Champaign (IL): National History Survey
Vassart M, Seguela A, Hayes H (1995) Chromosomal evolution in gazelles. J Hered 86:216–266
Veyrunes F, Catalan J, Sicard B, Robinson TJ, Duplantier JM, Granjon L, Dobigny G, Britton-Davidian J (2004) Autosome and sex chromosome diversity among the African pygmy mice, subgenus Nannomys (Murinae; Mus). Chromosome Res 12:369–382
White MJD (1978) Modes of speciation. WH Freeman, San Francisco
Acknowledgments
This work was supported by the project “CEITEC” Central European Institute of Technology (ED1.1.00/02.0068) from European Regional Development Funds, partly by Grant No. P502/11/0719 from the Grant Agency of the Czech Republic (HC, SK, OK, MV, JR) and by grants from the South African National Research Foundation (TJR, CAM).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
(a) Gel showing satellite DNA patterns after PCR analysis with SI (detection of satI DNA) and SII (detection of satII DNA) primers and dissected X;BTA5 fusion sites as templates. The PCR amplicons derived from NDR, GLE and ACE were prepared in Cernohorska et al. (2012) study. (b) Gel showing satellite DNA patterns after PCR analysis with SI and SII primers and dissected NDRX functional centromeres as template. PK = positive control, NK = negative control; molecular weights are represented on the first line (GIF 89 kb)
Table S1
List of chromosome fusions identified within Antilopini of the X;BTA5 clade by G- banding and comparative painting with whole-chromosome and region-specific probes derived from cattle. Classification of Antilopini was carried out following Groves and Grubb (2011). The highlighted characters were modified with regard to the FISH results published in Cernohorska et al. (2012) (see the text). R = References: 1 = Cernohorska et al. (2012); 2 = O´Brien et al. (2006); 3 = Vassart et al. (1995); 4 = Kumamoto et al. (1995). (PDF 40 kb)
Table S2
Presence and absence matrix of the chromosomal rearrangement included in this study. Taxon names are abbreviated to represent the genus and first two letters of the species in each instance. BTA, MKI and AMA represent outgroup taxa (see text for details). (PDF 38 kb)
Rights and permissions
About this article
Cite this article
Cernohorska, H., Kubickova, S., Kopecna, O. et al. Nanger, Eudorcas, Gazella, and Antilope form a well-supported chromosomal clade within Antilopini (Bovidae, Cetartiodactyla). Chromosoma 124, 235–247 (2015). https://doi.org/10.1007/s00412-014-0494-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-014-0494-5