Abstract
BnSIP1-1 is a trihelix transcription factor family gene which functions in abiotic stress response and abscisic acid (ABA) signaling during seed germination and seedling growth of Brassica napus. In the present study, further sequence analysis and phenotype identification indicated that this gene had roles in light regulation and flowering of reproductive growth stage. Many phytohormones responsive cis-acting elements, including TC-rich repeats, GARE-motif, and TCA and TGA elements, were identified in the promoter sequence of BnSIP1-1. The expression of BnSIP1-1 was regulated by light period and remarkable higher expression level of BnSIP1-1 was detected in roots than in leaves. Overexpression of BnSIP1-1 in Arabidopsis delayed flowering time for 3–5 days in transgenic plants. In addition, we also found BnSIP1-1 can respond to abiotic and ABA stress (treated with 200 mM NaCl, 300 mM mannitol or 50 μM ABA for 0, 1, 6, and 24 h) in B. napus through adjusting not only ABA but also other endogenous hormones, including indole-3-acetic acid and salicylic acid. Moreover, jasmonates (JA) signaling pathway was found not involving in the pathway of BnSIP1-1 responding to abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassica napus 6b INTERACTING PROTEIN1-1, BnSIP1-1, a SIP subfamily gene of trihelix transcription factor family, was recently characterized playing important roles in abiotic stress response and ABA signaling based on the previous work of our lab (Luo et al. 2017). BnSIP1-1 was also the first trihelix gene functionally identified in B. napus. The transcript level of BnSIP1-1 can be upregulated by drought, ionic toxicity, and ABA treatment. Overexpression of BnSIP1-1 could improve the germination rate of seeds under osmotic and salt stress as well as increasing the tolerance to osmotic stress during the seedling stage (Luo et al. 2017).
As we know, the genes of trihelix family were widely recognized to be involved in the regulation of light-responsive genes (Gilmartin et al. 1992; Kaplan-Levy et al. 2012). The family genes were first known as GT factors because they can bind GT elements of light-regulated genes, such as GT-1, GT-2, and GT-3a (Gilmartin et al. 1992; Hiratsuka et al. 1994; Ayadi et al. 2004). The core sequence of the GT element, 5′-G-Pu-(T/A)-A-A-(T/A)-3′, was sufficient for light induction and provided the factor’s name (Kaplan-Levy et al. 2012). More than 30 members of this family had been functionally revealed in different plant species. So far, there were more than 10 plant species in which the genomic-wide identification and expression profiling of trihelix family had been reported, including wheat, bamboo, cotton, rice, and oil crops (Ma et al. 2019; Cheng et al. 2019; Xiao et al. 2019; Mo et al. 2019; Li et al. 2019; Magwanga et al. 2019; Wang et al. 2016a, b, 2017, 2018, 2019; Song et al. 2016; Yu et al. 2015; Osorio et al. 2012; Fang et al. 2010; Qin et al. 2014).
Based on the evolution relationship and conserved domain, the family genes are classified into five clades, named GT-1, GT-2, SH4, SIP1, and GTγ. The function of the SIP1 clade has not been widely characterized. Only a few SIP1 members have been functionally identified such as ASIL1, ASIL2, FIP2, and AST1 (Gao et al. 2009; Barr et al. 2012; Geraldo et al. 2009; Xu et al. 2018). FIP2 was the first SIP clade gene discovered playing the indirect role in regulating other flowering-related genes, because it can interact tightly with FRIGIDA (FRI) in a yeast two hybrid screen (Geraldo et al. 2009). FRI and FLC together conferred a vernalization requirement for flowering. However, fip2 loss-of-function mutants did not affect the FLC expression which could be promoted by FRI (Geraldo et al. 2009), and the significance of the interaction remains uncertain.
Brassica Napus is a significant oil-producing crop, especially in China. Flowering is a complicated process that is affected by many internal and external factors. Control of flowering time of B. napus means a lot for crop production in different geographical regions. Many QTLs and candidates genes involving in flowering time variation have been reported (Raman et al. 2013; Wang et al. 2016a, b). A great number of homologues of Arabidopsis flowering genes such as FLC, FT, and CO have been identified as functionally conserved in B. napus (Tadege et al. 2001; Hou et al. 2012). However, the detailed molecular mechanism towards flowering regulation in B. napus is still unknown. Here, through further identifying the function of BnSIP1-1, we found it has a definite function of regulating flowering time. Overexpressing BnSIP1-1 in Arabidopsis can delay flowering time for 3–5 days. In addition, our data also indicated BnSIP1-1 was a clock gene which was regulated by light. However, ASIL1, the orthologue of this gene in Arabidopsis did not show a similar function in flowering control, suggesting functional differentiation of homologous genes in different species. In addition, we further confirmed this gene can adjust salicylic acid (SA) and indole-3-acetic acid (IAA) content in plant cells. IAA and SA content were increased in BnSIP1-1 overexpressing transgenic plants under normal growth condition, and had differential response to osmotic stress and ion toxicity. In summary, the present study provides more information on B. napus flowering pathways and sheds light on the possible relationship of abiotic stress, flowering control, and hormone equilibrium homeostasis.
Materials and Methods
Plant Materials, Growth Conditions
Wildtype Arabidopsis thaliana Columbia ecotype (Col-0) seeds were used in this study. The seeds (wildtype and transgenic plants) were sterilized in 2% NaClO with 0.02% Triton X-200 and planted on Murashige & Skoog (MS) medium, then vernalized in the darkness at 4 °C for 3 days. The seeds on MS medium were transferred to a culture room at 22 °C with a 16-h/8-h light/dark photoperiod. After 10 days, the seedlings were transferred to soil and placed in a growth chamber at 22 °C with 40–65% relative humidity and a 16-h light/8-h dark photoperiod.
The B. napus lines cv. ZhongShuang 6 (Short for ZS6), an elite Chinese cultivar in China and their transgenic plants were germinated in Petri dishes in the dark at 23 °C for 4 days. Seedlings were transferred to plastic plots filled with half-strength (1/2) Hoagland’s nutrient medium and grown in a growth chamber in a controlled environment (22 °C with 16-h light/8-h dark, 40–65% humidity, and light intensity of 8000 lx). All plant tissues used for further experiments were harvested and immediately frozen in liquid nitrogen after harvesting and stored at − 80 °C.
Isolation and Sequence Analysis of BnSIP1-1 from B. napus
The full-length BnSIP1-1 cDNA was isolated as previously described (Luo et al. 2017). The cDNA sequence was subjected to BLASTn against B. napus genome database at NCBI. The BnSIP1-1 promoter sequence was isolated using a Universal Genome Walker kit (Clontech) using ZS6 genomic DNA as templates. Putative functional cis elements in the promoter sequence were identified with PlantCARE software (bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plasmid Construction and Plant Transformation
The CDS of BnSIP1-1 was subcloned into the PBI121S vector under the cauliflower mosaic virus 35S promoter and a terminal poly A sequence. The 35S:BnSIP1 recombinant vector was introduced into Agrobacterium tumefaciens GV3101 by electroporation. A single positive colony was used to transform Arabidopsis (Col-0) and B. napus (ZS6). The transgenic Arabidopsis plants were generated by Agrobacterium-mediated floral dipping method (Clough and Bent 1998). The transgenic B. napus plants were generated by Agrobacterium-mediated method as reported previously (Luo et al. 2017). There were totally 17 independent T0 generation of transgenic Arabidopsis plants and 22 independent T0 generation of transgenic B. napus generated. Positive T3 generation of transgenic plants were grown for further experiments. The transgenic Arabidopsis lines OE-4, OE-5, and OE-9 were selected to represent lines with low, middle, and high expression level of BnSIP1-1. In addition, the transgenic B. napus line with highest expression level of BnSIP1-1 was used for detecting the IAA, JA, and SA Content.
RNA Extraction, cDNA Synthesis, and Transcription Analysis
For circadian rhythmicity expression detection of BnSIP1-1, the leaves and roots of 15-day-old B. napus seedlings grown in normal cultivation condition were collected for RNA isolation every 4 h, which was synchronal to the lighting. For detecting the expression of BnSIP1-1 in wildtype Arabidopsis and BnSIP1-1 overexpressing transgenic Arabidopsis, leaves of 4-week seedlings grown in normal cultivation condition were collected for RNA isolation. RNA extraction and the first-strand cDNA synthesis were performed using an RNAprep Pure Plant Kit (TIANGEN BIOTECH, Beijing, China) and Fast Quant RT Kit (with gDNase; TIANGENBIOTECH, Beijing, China) based on the manufacturers’ instructions. The concentration of RNA used for transcription in this study was about 100 ng/µl. The transcription analysis was performed using a SYBR Green kit (Bio-Rad, Hercules, CA, USA) on the CFX96 real-time PCR platform (Bio-Rad, Hercules, CA, USA). Three independent biological replicates were performed and the significance was determined through t test of SPSS statistical software (p < 0.05). The B. napus β-actin gene (accession No. AF111812.1) and Arabidopsis β-actin gene (accession No.AK317453.1) were used as internal reference controls, respectively. Relative expression levels were calculated by the 2−ΔΔCt method (Schmittgen and Livak 2008).
Determination of IAA, JA, and SA Content
For IAA, JA, and SA content measurement, 15-day-old BnSIP1-1 overexpression transgenic and wildtype B. napus those were cultivated in 1/2 Hoagland’s liquid nutrient medium were randomly transferred to plastic plots filled with control or stress treatment medium. ABA treatment was applied by adding 50 μM ABA to 1/2 Hoagland’s liquid medium. Salt or drought treatments were applied by adding 200 mM NaCl or 300 mM mannitol to 1/2 Hoagland’s liquid medium, respectively. Leaves were harvested at 0, 1, 6, and 24 h after each treatment. In order to study the roles of BnSIP1-1 in regulating hormone metabolism during seedling stage, we chose a very sensitive period when the first pair of true leaves was unfolded. These treatment doses and time points were applied according to our previous study (Luo et al. 2017). Quantification of endogenous IAA, JA, and SA was performed by using derivatization approach coupled with nano-LC–ESI-Q-TOF–MS analysis as described (Chen et al. 2012).
Results
Characterization of the 5′-Flanking Regulatory Region of BnSIP1-1
To investigate the transcriptional regulation of BnSIP1-1, 1429 bp 5′-flanking region upstream of the exon was isolated. There were 22 cis-acting elements (Table 1). By comparing the types and numbers of promoter elements, we found that there were many cis-acting elements with the same function. As we expected, several defense and phytohormone responsive cis-acting elements were identified, including TC-rich repeats, GARE-motif, and TCA and TGA elements. In addition, it is noteworthy that the promoter sequence contained a large number of light-responsive motifs, including Box-II, G-box, AE-box, GAG-motif, and as-2-box (Table 1). Circadian regulatory element is a very important motif for circadian control. The conserved sequence CAANNNNATC was founded at the 845 bp from the start codon of negative strand. The identification of light-responsive motifs and circadian regulatory elements can provide valuable hints for the potential relationship between BnSIP1-1 gene and light regulation.
Circadian Rhythmicity Regulation of Expression of BnSIP1-1
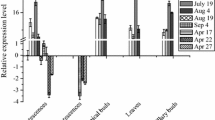
Based on the identification of several light-responsive elements and one circadian clock-involved element in promoter region of BnSIP1-1 gene, we supposed the expression of BnSIP1-1 was regulated by light. Therefore, we further tested the transcript level of BnSIP1-1 in leaves and roots using Real-time PCR at different time points. The result revealed that BnSIP1-1 transcript in leaves was sharply decreased after short-term exposure to light and gradually increased lately to peak level during the early dark (Fig. 1). Light inhibited the expression of BnSIP1-1 gene in leaves, making it reach the lowest expression value in the middle of the day and the highest expression in the middle of the night. However, notably, the BnSIP1-1 presented reverse rhythmic amplitude of expression in roots although the roots did not sense light at all. The transcript of BnSIP1-1 in roots was considerably increased when the circadian clock shifted from night to day, reaching transcription peak after 4 h of illumination. The peak level in roots was about 325 times higher than in leaves (Fig. 1). These data indicated BnSIP1-1 was a clock gene with reverse regulation mode in roots and leaves which was regulated by light.
Transcript quantitation of BnSIP1-1 during 24 h of light/dark circle. The leaves and roots of 15-day-old B. napus seedlings were collected every 4 h, which was synchronal to the lighting. The values were normalized to β-actin control. Values for each time point were means ± SD (n = 3). White bars: light on, dark bars: light off
Changes of IAA, JA, and SA Content in Transgenic Plants During Stress Conditions
Hormones are considered to be primary components of the signaling pathways those affect plant’ growth and development, as well as their adaptive response to environmental changes. Concentrations of IAA and SA in transgenic plants were significantly higher than those in wildtype plants under the CK treatment (Fig. 2). Under salt stress, the regulation mode of IAA in transgenic materials was completely opposite to wildtype (Fig. 2). However, when under ABA and mannitol treatments, the change pattern of IAA amount in transgenic plants were similar with in wildtype plants (Fig. 2). In the term of the concentration of SA, only at the beginning (1 h) of salt treatment the change pattern of SA amount was opposite to wildtype. After then, the change pattern in transgenic plants was similar with in wildtype plants. When under ABA and mannitol treatments, the change pattern of SA amount in transgenic plants were similar with in wildtype plants. Both the JA calculated amount and change pattern presented no significant difference between the transgenic and wildtype plants under NaCl, ABA, and mannitol treatments. These results indicated BnSIP1-1 regulated hormone metabolism as well as their response to abiotic stress and different hormones were regulated by different mode.
Changes of endogenous hormones content in transgenic plants during stress conditions. 15-day-old seedlings were transferred to 1/2 MS medium supplemented with 50 µM ABA, 200 mM NaCl, or 300 mM mannitol for 0 h, 1 h, 6 h, and 24 h. The concentration of endogenous hormones IAA, JA, and SA in leaves of seedlings was monitored. Data were means ± SD (n = 3) for each treatment. Significant differences from control plants are indicated by *p < 0.05 and **p < 0.01
Regulation of Flowering Time
Many light-responsive genes play roles in regulation of the flowering process, therefore we recorded the flowering time of overexpressing BnSIP1-1 transgenic Arabidopsis plants to further identify BnSIP1-1 function. We examined the flowering time of three independent lines in the homozygous T3 generation those were grown in greenhouse under long-days condition. The transgenic Arabidopsis plants bloomed 3–5 days later than wildtype Arabidopsis (Fig. 3). Moreover, the phenotypic alteration of flowering time in transgenic Arabidopsis was positively related to the expression of BnSIP1-1 in transgenic Arabidopsis. That meant the higher the expression of BnSIP1-1 in transgenic plants was, the longer the flowering time delayed. It indicated that BnSIP1-1 regulated flowering time in a dosage-dependent manner.
Comparative analysis of flowering time between BnSIP1-1 transgenic Arabidopsis and wildtype. (a) comparison of BnSIP1-1 transcript expression level and flowering days in wildtype Arabidopsis and BnSIP1-1 overexpressing transgenic plants. Data were means ± SD of three biological replicates. The asterisks * and ** indicated that the value of a t test was p < 0.05 and p < 0.01, respectively, and a significant difference between the different transgenic lines and wildtype. b Morphological comparison of flowering time between BnSIP1-1 overexpressing transgenic plants and wildtype plants
Discussion
Recently, BnSIP1-1 was identified as a trihelix family gene mediating abiotic stress response and ABA signaling in B. napus (Luo et al. 2017). Overexpressing BnSIP1-1 could increase germination ability of seeds under drought, salt, and ABA treatment, as well as improving osmotic stress tolerance and ABA sensitivity during early stage of vegetative growth (Luo et al. 2017). This remarkable phenotype prompted us to explore the function of the gene in more depth. Through further study, we found additional pathways that BnSIP1-1 is involved in, including photoperiod, flowering time, and hormone regulation.
Circadian Clock Gene BnSIP1-1 Expressing Higher in Roots Than in Leaves
Based on the 24-h expression level monitoring, we found the transcript abundance of BnSIP1-1 in leaves and roots both were affected by photoperiod. As dawn approached, BnSIP1-1 transcript in leaves decayed and reached the lowest point after 4 h of light (Fig. 1). Certainly, we cannot exclude the possibility that lower expression level may be detected if time points during 4 h to 8 h of light period were adopted to monitor. However, this result at least demonstrated that light repressed the transcription of BnSIP1-1 in leaves. This repression was not persistent because the expression level slowly rose when morning came and rhythmically reached peak before night in leaves. On the contrary, BnSIP1-1 was expressed at peak levels after 4 h of light and sharply decreased after following 4 h of light. Moreover, the expression level in roots was notably higher in roots than in leaves. It seemed light regulated the expression of BnSIP1-1 more strongly in root than in leaves, although leaves were recognized as plant tissue that received light (Endo et al. 2005; Łabuz et al. 2012). Actually, although roots are buried in soil, it can also perceive light signal directly by sensing light through root photoreceptors (Lee et al. 2016; Suzuki et al. 2011; Sassi et al. 2012) and directly and indirectly transporting of light signal by mobile signaling messengers ( Lee et al. 2017; Saini et al. 2013; Rakusová et al. 2016; Xu et al. 2016). We deduced BnSIP1-1 was a potential root photoreceptor or regulated by mobile light signaling messengers.
BnSIP1-1 Response to Abiotic and ABA Stress Through Adjusting Multiple Endogenous Hormones
ABA, IAA, JA, and SA are known to play major roles in mediating plant defense response against biotic and abiotic stresses (Bari and Jones 2009; Nakashima and Yamaguchi-Shinozaki 2013). A previous study indicated BnSIP1-1 changed ABA accumulation (Luo et al. 2017), therefore three other significant hormones IAA, JA, and SA were also monitored under stress treatments at this study. IAA is a key hormone promoting plant growth. IAA synthesis will be decreased and plant growth will be inhibited when plant cell encounters drought (Dong et al. 2019). In addition, osmotic stress induces increased endogenous SA amount in many plants (Munn-Bosch and Penuelas 2003; Miura and Tada 2014). SA modulates plant abiotic stress through different pathways such as stomatal conductance, antioxidant defense system, and NO production (Khokon et al. 2011; Saruhan et al. 2012; Hayat et al. 2008). IAA and SA content were increased in BnSIP1-1 overexpressing transgenic plants, indicating BnSIP1-1 played a critical role in controlling endogenous IAA and SA accumulation to response to abiotic stresses. Under ABA and mannitol treatment, the change patterns of IAA and SA content were similar along with the time extension of stress treatment time between transgenic plants and wildtype. Conversely, under salt stress treatment, the change patterns of IAA and SA content were different between transgenic plants and wildtype. Although the content of IAA and SA was higher in the transgenic plants than in wildtype without any stress treatment, after 24 h of salt treatment, the content of IAA and SA decreased to the similar level as wildtype, which was consistent with the phenotype that transgenic plant cannot survive when treated with high concentration of salt as previously reported (Luo et al. 2017). It indicated that IAA and SA took part in the regulation pathway of BnSIP1-1 responding to drought and ABA stress, but not salt stress. JA mainly participates in the defense against insect attack and wounding (Wasternack and Song 2017), but recently some researchers reported it can also play roles in osmotic stress or salt stress (Chen et al. 2018; Toda et al. 2013). GmSK1-overexpressing transgenic tobacco plants showed enhanced tolerance to high salinity and drought stress and its expression level can be induced by JA (Chen et al. 2018). Toda reported that RICE SALT SENSITIVE3 (RSS3) regulated root growth under salt stress and the expression of a significant portion of JA-responsive genes was upregulated in rss3 mutant (Toda et al. 2013), suggesting that JA played potential important roles in plant tolerance to abiotic stresses. However, in our study, there was not any difference in the content of JA under normal growth condition and stresses treatments between transgenic plants and wildtype. Moreover, the pattern of change in JA content was also similar between transgenic plants and wildtype. The results presented here suggested that JA did not participate in the regulation of the BnSIP1-1-related abiotic response-signaling pathway.
BnSIP1-1 Can Regulate Flowering Time
Transgenic Arabidopsis overexpressing BnSIP1-1 flowered later compared with wildtype. This is the first SIP1 Clade gene of the trihelix family to be characterized with an effect on flowering. The homologue of BnSIP1-1 in Arabidopsis is ASIL1, which does not involve in flowering but is a regulator that represses the expression of embryonic seed maturation genes in vegetative tissues (Gao et al. 2009). Although another SIP1 clade gene FIP2 can interact with FRIGIDA (FRI), the loss-of-function mutant did not show phenotype regarding flowering times (Geraldo et al. 2009) and there are no reports on the phenotype of overexpressing plants. It is noteworthy that BnSIP1-1 affected flowering in a dosage-dependent manner. Whether this manner of regulation is related to the levels of ABA/SA/IAA hormone is still unknown and needs further study. In our previous study, BnSIP1-1 could promote the expression level of BnNAC45, however, the phenotype of overexpressing transgenic plants was not consistent totally because they both can increase the tolerance of plants to osmotic and ionic stress but BnNAC45 OE (overexpressing) plants had ABA hyposensitivity phenotype, while BnSIP1-1 OE plants had ABA-tolerant phenotype (Ying et al. 2014; Luo et al. 2017). In this study, we found another inconsistent phenotype. BnNAC45 OE transgenic Arabidopsis bloomed about 2 days earlier than wildtype (Ying et al. 2014), while BnSIP1-1 OE transgenic plants bloomed 3–5 days later than wildtype. We speculated BnSIP1-1 and BnNAC45 both were involved in the ABA-dependent abiotic stress response pathway and regulated plants flowering time through their circadian clock characteristic but they were in different feedback regulation nodes of this complex signaling network. Future research should focus on the characterization of these genes as well as their relationship with hormone equilibrium homeostasis.
In conclusion, we present evidence that an abiotic response gene, BnSIP1-1, was also involved in light rhythm regulation and flowering of reproductive growth stage in B. napus. Light inhibited the expression of BnSIP1-1 gene in leaves but stimulated its expression in roots in B. napus, indicating this gene may be a potential root photoreceptor or regulated by mobile light signaling messengers. In addition, this gene can regulate IAA and SA, but not JA homostasis. Future study may provide additional insights into the mechanism by which BnSIP1-1 exerts its function on both flowering and abiotic stress tolerance. Whether the later flowering and abiotic stress-resistant phenotype of BnSIP1-1 overexpressing plants have direct relationship with hormone signal transduction and light regulation? Our study will provide better understanding of the molecular functions of BnSIP1-1 in plants.
References
Ayadi M, Delaporte V, Li YF, Zhou DX (2004) Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett 562:147–154
Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488
Barr MS, Willmann MR, Jenik PD (2012) Is there a role for trihelix transcription factors in embryo maturation? Plant Signal Behav 7:205–209
Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A (2001) The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11:731–753
Chen M, Fu X, Liu J, Ye T, Hou S, Huang Y et al (2012) Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC–ESI-Q-TOF-MS analysis. J Chromatogr B 905:67–74
Chen Y, Chi Y, Meng Q, Wang X, Yu D (2018) GmSK1, an SKP1 homologue in soybean, is involved in the tolerance to salt and drought. Plant Physiol Biochem 127:25–31
Cheng X, Xiong R, Yan H, Gao Y, Liu H, Wu M, Xiang Y (2019) The trihelix family of transcription factors: functional and evolutionary analysis in Moso bamboo (Phyllostachys edulis). BMC Plant Biol 19:154
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Conley TR, Park SC, Kwon HB, Peng HP, Shih MC (1994) Characterization of cis-acting elements in light regulation of the nuclear gene encoding the A subunit of chloroplast isozymes of glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. Mol Cell Biol 14:2525–2533
Diaz-De-Leon F, Klotz KL, Lagrimini LM (1993) Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene. Plant Physiol 101:1117–1118
Dong S, Jiang Y, Dong Y, Wang L, Wang W, Ma Z, Yan C, Ma C, Liu L (2019) A study on soybean responses to drought stress and rehydration. Saudi J Biol Sci 26:2006–2017
Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A (2005) Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell 17:1941–1952
Fang Y, Xie K, Hou X, Hu H, Xiong L (2010) Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol Genet Genom 283:157–169
Feldbrügge M, Sprenger M, Dinkelbach M, Yazaki K, Harter K, Weisshaar B (1994) Functional analysis of a light-responsive plant bZIP transcriptional regulator. Plant Cell 6(11):1607–1621
Gao M-J, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD et al (2009) Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21:54–71
Geraldo N, Bäurle I, Kidou S, Hu X, Dean C (2009) FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150:1611–1618
Gilmartin PM, Memelink J, Hiratsuka K, Kay SA, Chua NH (1992) Characterization of a gene encoding DNA binding protein with specificity for a light-responsive element. Plant Cell 4:839–849
Hayat S, Hasan SA, Fariduddin Q, Ahmad A et al (2008) Growth of tomato (Lycopersicon esculentum) in response to salicylic acid under water stress. J Plant Interact 3:297–304
Hennig J, Dewey RE, Cutt JR, Klessig DF (1993) Pathogen, salicylic acid and developmental dependent expression of a beta-1,3-glucanase/GUS gene fusion in transgenic tobacco plants. Plant J 4:481–493
Hiratsuka K, Wu X, Fukuzawa H, Chua NH (1994) Molecular dissection of GT-1 from Arabidopsis. Plant Cell 6:1805–1813
Hou J, Long Y, Raman H, Zou X, Wang J, Dai S, Xiao Q, Li C, Fan L, Liu B et al (2012) A tourist-like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed (Brassica Napus L.). BMC Plant Biol 12:238
Kaplan-Levy RN, Brewer PB, Quon T, Smyth DR (2012) The trihelix family of transcription factors–light, stress and development. Trends Plant Sci 17:163–171
Khokon AR, Okuma E, Hossain MA, Munemasa S, Uraji MN (2011) Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ 34:434–443
Łabuz J, Sztatelman O, Banaś AK, Gabryś H (2012) The expression of phototropins in Arabidopsis leaves: developmental and light regulation. J Exp Bot 63:1763–1771
Lee HJ, Ha JH, Kim SG, Choi HK, Kim ZH, Han YJ (2016) Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots. Sci Signal 9:106
Lee HJ, Park YJ, Ha JH, Baldwin IT, Park CM (2017) Multiple routes of light signaling during root photomorphogenesis. Trends Plant Sci 22:803–812
Li J, Zhang M, Sun J, Mao X, Wang J, Wang J, Liu H, Zheng H, Zhen Z, Zhao H, Zou D (2019) Genome-wide characterization and identification of trihelix transcription factor and expression profiling in response to abiotic stresses in rice (Oryza sativa L.). Int J Mol Sci 20:251
Luo J, Tang S, Mei F, Peng X, Li J, Li X et al (2017) BnSIP1-1, a trihelix family gene, mediates abiotic stress tolerance and ABA signaling in Brassica napus. Front Plant Sci 8:44
Magwanga RO, Kirungu JN, Lu P, Yang X, Dong Q, Cai X, Xu Y, Wang X, Zhou Z, Hou Y, Nyunja R, Agong SG, Hua J, Zhang B, Wang K, Liu F (2019) Genome wide identification of the trihelix transcription factors and overexpression of Gh_A05G2067 (GT-2), a novel gene contributing to increased drought and salt stresses tolerance in cotton. Physiol Plant 167:447–464
Ma Z, Liu M, Sun W, Huang L, Wu Q, Bu T, Li C, Chen H (2019) Genome-wide identification and expression analysis of the trihelix transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol 19:344
Miura K, Tada Y (2014) Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci 5
Mo H, Wang L, Ma S, Yu D, Lu L, Yang Z, Yang Z, Li F (2019) Transcriptome profiling of Gossypium arboreum during fiber initiation and the genome-wide identification of trihelix transcription factors. Gene 709:36–47
Munn-Bosch S, Peuelas J (2003) Photo- and antioxidative protection, and a role for salicylic acid during drought and recovery in field-grown Phillyrea angustifolia plants. Planta 217(5):758–766
Nakashima K, Yamaguchi-Shinozaki K (2013) ABA signaling in stress-response and seed development. Plant Cell Rep 32:959–970
Nash J, Luehrsen KR, Walbot V (1990) Bronze-2 gene of maize: reconstruction of a wild-type allele and analysis of transcription and splicing. Plant Cell 2:1039–1049
Osorio MB, Bücker-Neto L, Castilhos G, Turchetto-Zolet AC, Wiebke-Strohm B, Bodanese-Zanettini MH, Margis-Pinheiro M (2012) Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet Mol Biol 35:233–246
Pastuglia M, Roby D, Dumas C, Cock JM (1997) Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9:49–60
Pichersky E, Bernatzky R, Tanksley SD, Breidenbach RB, Kausch AP, Cashmore AR (1985) Molecular characterization and genetic mapping of two clusters of genes encoding chlorophyll a/b-binding proteins in Lycopersicon esculentum (tomato). Gene 40:247–258
Qin Y, Ma X, Yu G, Wang Q, Wang L, Kong L, Kim W, Wang HW (2014) Evolutionary history of trihelix family and their functional diversification. DNA Res 21:499–510
Rakusová H, Abbas M, Han H, Song S, Robert HS, Friml J (2016) Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr Biol 26:3026–3032
Raman H, Raman R, Eckermann P, Coombes N, Manoli S, Zou X, Edwards D, Meng J, Prangnell R, Stiller J et al (2013) Genetic and physical mapping of flowering time loci in canola (Brassica Napus L.). Theor Appl Genet 126:119–132
Rundle SJ, Zielinski RE (1991) Organization and expression of two tandemly oriented genes encoding ribulosebisphosphate carboxylase/oxygenase activase in barley. J Biol Chem 266:4677–4685
Saini S, Sharma I, Kaur N, Pati PK (2013) Auxin: a master regulator in plant root development. Plant Cell Rep 32:741–757
Saruhan N, Saglam A, Kadioglu A (2012) Salicylic acid pretreatment induces drought tolerance and delays leaf rolling by inducing antioxidant systems in maize genotypes. Acta Physiol Plant 34:97–106
Sassi M, Lu Y, Zhang Y, Wang J, Dhonukshe P, Blilou I (2012) COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1- and PIN2-dependent auxin transport in Arabidopsis. Development 139:3402–3412
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108
Song A, Wu D, Fan Q, Tian C, Chen S, Guan Z, Xin J, Zhao K, Chen F (2016) Transcriptome-wide identification and expression profiling analysis of chrysanthemum trihelix transcription factors. Int J Mol Sci 17:198
Suzuki A, Suriyagoda L, Shigeyama T, Tominaga A, Sasaki M, Hiratsuka Y et al (2011) Lotus japonicus nodulation is photomorphogenetically controlled by sensing the red/far red (R/FR) ratio through jasmonic acid (JA) signaling. Proc Natl Acad Sci 108:16837–16842
Tadege M, Sheldon CC, Helliwell CA, Stoutjesdijk P, Dennis ES, Peacock WJ (2001) Control of flowering time by FLC orthologues in Brassica Napus. Plant J 28:545–553
Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y et al (2013) RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell 25:1709–1725
Wang N, Chen B, Xu K, Gao G, Li F, Qiao J, Yan G, Li J, Li H, Wu X (2016a) Association mapping of flowering time QTLs and insight into their contributions to rapeseed growth habits. Front Plant Sci 7:338
Wang Z, Liu Q, Wang H, Zhang H, Xu X, Li C, Yang C (2016b) Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Sci Rep 6:36274
Wang Z, Zhao K, Pan Y, Wang J, Song X, Ge W, Yuan M, Lei T, Wang L, Zhang L, Li Y, Liu T, Chen W, Meng W, Sun C, Cui X, Bai Y, Wang X (2018) Genomic, expressional, protein-protein interactional analysis of Trihelix transcription factor genes in Setaria italia and inference of their evolutionary trajectory. BMC Genom 19:665
Wang C, Wang Y, Pan Q, Chen S, Feng C, Hai J, Li H (2019) Comparison of Trihelix transcription factors between wheat and Brachypodium distachyon at genome-wide. BMC Genom 20:142
Wang W, Wu P, Liu TK, Ren H, Li Y, Hou X (2017) Genome-wide analysis and expression divergence of the trihelix family in brassica rapa: Insight into the evolutionary patterns in plants. Sci Rep 7(1)
Wasternack C, Song S (2017) Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J Exp Bot 68:1303–1321
Werneke JM, Chatfield JM, Ogren WL (1989) Alternative mRNA splicing generates the two ribulosebisphosphate carboxylase/oxygenase activase polypeptides in spinach and Arabidopsis. Plant Cell 1:815–825
Xiao J, Hu R, Gu T, Han J, Qiu D, Su P, Feng J, Chang J, Yang G, He G (2019) Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat. BMC Genom 20:287
Xu D, Jiang Y, Li J, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci 113:7655–7660
Xu H, Shi X, He L, Guo Y, Zang D, Li H, Zhang W, Wang Y (2018) Arabidopsis thaliana trihelix transcription factor AST1 mediates salt and osmotic stress tolerance by binding to a novel AGAG-box and some GT motifs. Plant Cell Physiol 59:946–965
Ying L, Chen H, Cai W (2014) BnNAC485 is involved in abiotic stress responses and flowering time in Brassica napus. Plant Physiol Biochem 79:77–87
Yu C, Cai X, Ye Z, Li H (2015) Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem Biophys Res Commun 468:653–659
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Number 31201152 to JL and Grant Number 31671733 to XY)
Author information
Authors and Affiliations
Contributions
JL conducted experimental design, data analysis, and paper writing; WJ conducted RT-PCR and hormone content analysis; ST conducted the plant transformation; FM assisted data analysis; XZ and XY revised the manuscript; GW designed the research.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Luo, J., Jiang, W., Tang, S. et al. BnSIP1-1 Involves in Light Response and Regulation of Endogenous Hormones and Flowering Time of Brassica Napus. J Plant Growth Regul 40, 2049–2057 (2021). https://doi.org/10.1007/s00344-020-10250-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-020-10250-w