Abstract
Objectives
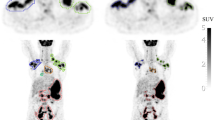
To demonstrate the effectiveness of automatic segmentation of diffuse large B-cell lymphoma (DLBCL) in 3D FDG-PET scans using a deep learning approach and validate its value in prognosis in an external validation cohort.
Methods
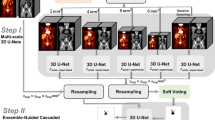
Two PET datasets were retrospectively analysed: 297 patients from a local centre for training and 117 patients from an external centre for validation. A 3D U-Net architecture was trained on patches randomly sampled within the PET images. Segmentation performance was evaluated by six metrics, including the Dice similarity coefficient (DSC), Jaccard similarity coefficient (JSC), sensitivity (Se), positive predictive value (PPV), Hausdorff distance 95 (HD 95), and average symmetric surface distance (ASSD). Finally, the prognostic value of predictive total metabolic tumour volume (pTMTV) was validated in real clinical applications.
Results
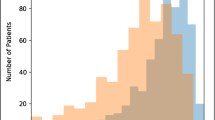
The mean DSC, JSC, Se, PPV, HD 95, and ASSD (with standard deviation) for the validation cohort were 0.78 ± 0.25, 0.69 ± 0.26, 0.81 ± 0.27, 0.82 ± 0.25, 24.58 ± 35.18, and 4.46 ± 8.92, respectively. The mean ground truth TMTV (gtTMTV) and pTMTV were 276.6 ± 393.5 cm3 and 301.9 ± 510.5 cm3 in the validation cohort, respectively. Perfect homogeneity in the Bland–Altman analysis and a strong positive correlation in the linear regression analysis (R2 linear = 0.874, p < 0.001) were demonstrated between gtTMTV and pTMTV. pTMTV (≥ 201.2 cm3) (PFS: HR = 3.097, p = 0.001; OS: HR = 6.601, p < 0.001) was shown to be an independent factor of PFS and OS.
Conclusions
The FCN model with a U-Net architecture can accurately segment lymphoma lesions and allow fully automatic assessment of TMTV on PET scans for DLBCL patients. Furthermore, pTMTV is an independent prognostic factor of survival in DLBCL patients.
Key Points
•The segmentation model based on a U-Net architecture shows high performance in the segmentation of DLBCL patients on FDG-PET images.
•The proposed method can provide quantitative information as a predictive TMTV for predicting the prognosis of DLBCL patients.





Similar content being viewed by others
Abbreviations
- ASSD:
-
Average symmetric surface distance
- DLBCL:
-
Diffuse large B-cell lymphoma
- DSC:
-
Dice similarity coefficient
- FCN:
-
Fully convolutional neural network
- gtTMTV:
-
Ground truth total metabolic tumour volume
- HD 95:
-
Hausdorff distance 95
- IQR:
-
Interquartile range
- JSC:
-
Jaccard similarity coefficient
- NCCN-IPI:
-
National Comprehensive Cancer Network International Prognostic Index
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- PPV:
-
Positive predictive value
- pTMTV:
-
Predictive total metabolic tumour volume
- ReLU:
-
Rectified linear unit
- ROC:
-
Receiver operating characteristic
- SD:
-
Standard deviation
- Se:
-
Sensitivity
- SUV:
-
Standardised uptake value
- TLG:
-
Total lesion glycolysis
- TMTV:
-
Total metabolic tumour volume
- VOI:
-
Volume of interest
References
Sehn LH, Salles G (2021) Diffuse large B-cell lymphoma. N Engl J Med 384:842–858
Camicia R, Winkler HC, Hassa PO (2015) Novel drug targets for personalized precision medicine in relapsed/refractory diffuse large B-cell lymphoma: a comprehensive review. Mol Cancer 14:207
International Non-Hodgkin’s Lymphoma Prognostic Factors Project (1993) A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med 329:987–994
Zhou Z, Sehn LH, Rademaker AW et al (2014) An enhanced international prognostic index (NCCN-IPI) for patients with diffuse large B-cell lymphoma treated in the rituximab era. Blood 124:982–982
Ruppert AS, Dixon JG, Salles G et al (2020) International prognostic indices in diffuse large B-cell lymphoma: a comparison of IPI, R-IPI, and NCCN-IPI. Blood 135:2041–2048
Sasanelli M, Meignan M, Haioun C et al (2014) Pretherapy metabolic tumour volume is an independent predictor of outcome in patients with diffuse large B-cell lymphoma. Eur J Nucl Med Mol Imaging 41:2017–2022
Mikhaeel NG, Smith D, Dunn JT et al (2016) Combination of baseline metabolic tumour volume and early response on PET/CT improves progression-free survival prediction in DLBCL. Eur J Nucl Med Mol Imaging 43:1209–1219
Jiang C, Teng Y, Zheng Z, Zhou Z, Xu J (2021) Value of total lesion glycolysis and cell-of-origin subtypes for prognostic stratification of diffuse large B-cell lymphoma patients. Quant Imaging Med Surg 11:2509–2520
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A (eds) Medical image computing and computer-assisted intervention – MICCAI 2015. MICCAI 2015. Lecture notes in computer science, vol 9351. Springer, Cham, pp 234–241
Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH (2020) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211
Weston AD, Korfiatis P, Kline TL et al (2019) Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 290:669–679
Ye Y, Cai Z, Huang B et al (2020) Fully-automated segmentation of nasopharyngeal carcinoma on dual-sequence MRI using convolutional neural networks. Front Oncol 10:166
Capobianco N, Meignan M, Cottereau A-S et al (2021) Deep-learning 18F-FDG uptake classification enables total metabolic tumor volume estimation in diffuse large B-cell lymphoma. J Nucl Med 62:30–36
Blanc-Durand P, Jégou S, Kanoun S et al (2021) Fully automatic segmentation of diffuse large B cell lymphoma lesions on 3D FDG-PET/CT for total metabolic tumour volume prediction using a convolutional neural network. Eur J Nucl Med Mol Imaging 48:1362–1370
Nioche C, Orlhac F, Boughdad S et al (2018) LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 78:4786–4789
Maas AL, Hannun AY, Ng AY (2013) Rectifier nonlinearities improve neural network acoustic models. Proc ICML 30:3
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell 40:834–848
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) The importance of skip connections in biomedical image segmentation. In: Carneiro G, Mateus D, Pete L (eds) Deep learning and data labeling for medical applications. Springer, Cham, pp 179–187
Shen D, Wu G, Suk H-I (2017) Deep learning in medical image analysis. Annu Rev Biomed Eng 19:221–248
Litjens G, Kooi T, Bejnordi BE et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Zhong Z, Kim Y, Zhou L et al (2018) 3D fully convolutional networks for co-segmentation of tumors on PET-CT images. Proc IEEE Int Symp Biomed Imaging 2018:228–231
Naser MA, van Dijk LV, He R, Wahid KA, Fuller CD (2020) Tumor segmentation in patients with head and neck cancers using deep learning based-on multi-modality PET/CT images. Head Neck Tumor Segmentation 12603:85–98
Zucca E, Cascione L, Ruberto T et al (2020) Prognostic models integrating quantitative parameters from baseline and interim positron emission computed tomography in patients with diffuse large B-cell lymphoma: post-hoc analysis from the SAKK38/07 clinical trial. Hematol Oncol 38:715–725
Chang C-C, Cho S-F, Chuang Y-W et al (2017) Prognostic significance of total metabolic tumor volume on (18)F-fluorodeoxyglucose positron emission tomography/ computed tomography in patients with diffuse large B-cell lymphoma receiving rituximab-containing chemotherapy. Oncotarget 8:99587–99600
Shagera QA, Cheon GJ, Koh Y et al (2019) Prognostic value of metabolic tumour volume on baseline 18F-FDG PET/CT in addition to NCCN-IPI in patients with diffuse large B-cell lymphoma: further stratification of the group with a high-risk NCCN-IPI. Eur J Nucl Med Mol Imaging 46:1417–1427
Vercellino L, Cottereau A-S, Casasnovas O et al (2021) High total metabolic tumor volume at baseline predicts survival independent of response to therapy. Blood 135:1396–1405
Zhao P, Yu T, Pan Z (2021) Prognostic value of the baseline 18F-FDG PET/CT metabolic tumour volume (MTV) and further stratification in low-intermediate (L-I) and high-intermediate (H-I) risk NCCNIPI subgroup by MTV in DLBCL MTV predict prognosis in DLBCL. Ann Nucl Med 35:24–30
Meignan M, Sasanelli M, Casasnovas RO et al (2014) Metabolic tumour volumes measured at staging in lymphoma: methodological evaluation on phantom experiments and patients. Eur J Nucl Med Mol Imaging 41:1113–1122
Funding
This work was supported in part by the National Nature Science Foundation of China under grant no. 62106101. This work was also supported in part by the Natural Science Foundation of Jiangsu Province under grant no. BK20210180. This work was also supported in part by the grant from Jiangsu Provincial Key R&D Program under no. BE2020620 and BE2020723.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jian He.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
•retrospective
•diagnostic and prognostic study
•performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chong Jiang and Kai Chen contributed to the work equally and should be regarded as co-first authors
Supplementary Information
ESM 1
(DOC 926 kb)
Rights and permissions
About this article
Cite this article
Jiang, C., Chen, K., Teng, Y. et al. Deep learning–based tumour segmentation and total metabolic tumour volume prediction in the prognosis of diffuse large B-cell lymphoma patients in 3D FDG-PET images. Eur Radiol 32, 4801–4812 (2022). https://doi.org/10.1007/s00330-022-08573-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08573-1