Abstract
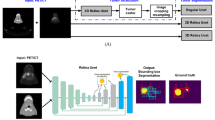
Segmentation of head and neck cancer (HNC) primary tumors on medical images is an essential, yet labor-intensive, aspect of radiotherapy. PET/CT imaging offers a unique ability to capture metabolic and anatomic information, which is invaluable for tumor detection and border definition. An automatic segmentation tool that could leverage the dual streams of information from PET and CT imaging simultaneously, could substantially propel HNC radiotherapy workflows forward. Herein, we leverage a multi-institutional PET/CT dataset of 201 HNC patients, as part of the MICCAI segmentation challenge, to develop novel deep learning architectures for primary tumor auto-segmentation for HNC patients. We preprocess PET/CT images by normalizing intensities and applying data augmentation to mitigate overfitting. Both 2D and 3D convolutional neural networks based on the U-net architecture, which were optimized with a model loss function based on a combination of dice similarity coefficient (DSC) and binary cross entropy, were implemented. The median and mean DSC values comparing the predicted tumor segmentation with the ground truth achieved by the models through 5-fold cross validation are 0.79 and 0.69 for the 3D model, respectively, and 0.79 and 0.67 for the 2D model, respectively. These promising results show potential to provide an automatic, accurate, and efficient approach for primary tumor auto-segmentation to improve the clinical practice of HNC treatment.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Siegel, R.L., Miller, K.D., Jemal, A.: Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30 (2020). https://doi.org/10.3322/caac.21590
Rosenthal, D.I., et al.: Beam path toxicities to non-target structures during intensity-modulated radiation therapy for head and neck cancer. Int. J. Radiat. Oncol. Biol. Phys. 72, 747–755 (2008). https://doi.org/10.1016/j.ijrobp.2008.01.012
Vorwerk, H., et al.: Protection of quality and innovation in radiation oncology: the prospective multicenter trial the German Society of Radiation Oncology (DEGRO-QUIRO study). Strahlentherapie und Onkol. 190, 433–443 (2014)
Riegel, A.C., et al.: Variability of gross tumor volume delineation in head-and-neck cancer using CT and PET/CT fusion. Int. J. Radiat. Oncol. Biol. Phys. 65, 726–732 (2006). https://doi.org/10.1016/j.ijrobp.2006.01.014
Rasch, C., Steenbakkers, R., Van Herk, M.: Target definition in prostate, head, and neck. Semin. Radiat. Oncol. 15, 136–145 (2005). https://doi.org/10.1016/j.semradonc.2005.01.005
Breen, S.L., et al.: Intraobserver and interobserver variability in GTV delineation on FDG-PET-CT images of head and neck cancers. Int. J. Radiat. Oncol. Biol. Phys. 68, 763–770 (2007). https://doi.org/10.1016/j.ijrobp.2006.12.039
Segedin, B., Petric, P.: Uncertainties in target volume delineation in radiotherapy–are they relevant and what can we do about them? Radiol. Oncol. 50, 254–262 (2016)
Anderson, C.M., et al.: Interobserver and intermodality variability in GTV delineation on simulation CT, FDG-PET, and MR images of head and neck cancer. Jacobs J. Radiat. Oncol. 1, 6 (2014)
Guo, Y., Liu, Yu., Georgiou, T., Lew, M.S.: A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 7(2), 87–93 (2017). https://doi.org/10.1007/s13735-017-0141-z
Garcia-Garcia, A., Orts-Escolano, S., Oprea, S., Villena-Martinez, V., Garcia-Rodriguez, J.: A Review on Deep Learning Techniques Applied to Semantic Segmentation (2017)
Boldrini, L., Bibault, J.-E., Masciocchi, C., Shen, Y., Bittner, M.-I.: Deep learning: a review for the radiation oncologist. Front. Oncol. 9, 977 (2019)
Cardenas, C.E., Yang, J., Anderson, B.M., Court, L.E., Brock, K.B.: Advances in auto-segmentation. Semin. Radiat. Oncol. 29, 185–197 (2019). https://doi.org/10.1016/j.semradonc.2019.02.001
Oreiller, V.A.V., Vallieres, M., Castelli, J., Boughdad, H.E.M.J.S., Adrien, J.O.P.: Automatic Segmentation of Head and Neck Tumors and Nodal Metastases in PET-CT scans (2020). http://proceedings.mlr.press/v121/andrearczyk20a.html
Li, L., Zhao, X., Lu, W., Tan, S.: Deep learning for variational multimodality tumor segmentation in PET/CT. Neurocomputing 392, 277–295 (2020)
Leung, K.H., et al.: A physics-guided modular deep-learning based automated framework for tumor segmentation in PET images. arXiv Preprint arXiv:2002.07969 (2020)
Kawauchi, K., et al.: A convolutional neural network-based system to classify patients using FDG PET/CT examinations. BMC Cancer 20, 1–10 (2020). https://doi.org/10.1186/s12885-020-6694-x
Zhong, Z., et al.: 3D fully convolutional networks for co-segmentation of tumors on PET-CT images. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 228–231. IEEE (2018)
Jemaa, S., Fredrickson, J., Carano, R.A.D., Nielsen, T., de Crespigny, A., Bengtsson, T.: Tumor segmentation and feature extraction from whole-body FDG-PET/CT using cascaded 2D and 3D convolutional neural networks. J. Digit. Imaging. 33, 888–894 (2020). https://doi.org/10.1007/s10278-020-00341-1
Zhao, X., Li, L., Lu, W., Tan, S.: Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys. Med. Biol. 64, 15011 (2018)
Huang, B., et al.: Fully automated delineation of gross tumor volume for head and neck cancer on PET-CT using deep learning: a dual-center study. Contrast Media Mol. Imaging 2018, 1–12 (2018). https://pubmed.ncbi.nlm.nih.gov/30473644/
Moe, Y.M., et al.: Deep learning for automatic tumour segmentation in PET/CT images of patients with head and neck cancers. arXiv Preprint arXiv:1908.00841 (2019)
Guo, Z., Li, X., Huang, H., Guo, N., Li, Q.: Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans. Radiat. Plasma Med. Sci. 3, 162–169 (2019)
Jin, D., et al.: Accurate esophageal gross tumor volume segmentation in PET/CT using two-stream chained 3D deep network fusion. In: Shen, D., Liu, T., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 182–191. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_21
Zhou, T., Ruan, S., Canu, S.: A review: deep learning for medical image segmentation using multi-modality fusion. Array 3, 100004 (2019)
AIcrowd MICCAI 2020: HECKTOR Challenges. https://www.aicrowd.com/challenges/miccai-2020-hecktor. Accessed 07 Sept 2020
Andrearczyk, V., et al.: Overview of the HECKTOR challenge at MICCAI 2020: automatic head and neck tumor segmentation in PET/CT. In: Andrearczyk, V., et al. (eds.) HECKTOR 2020. LNCS, vol. 12603, pp. 1–21. Springer, Cham (2021)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Naser, M.A., Deen, M.J.: Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med. 121, 103758 (2020). https://doi.org/10.1016/j.compbiomed.2020.103758
Springenberg, J.T., Dosovitskiy, A., Brox, T., Riedmiller, M.: Striving for simplicity: the all convolutional net. arXiv Preprint arXiv:1412.6806 (2014)
Noh, H., Hong, S., Han, B.: Learning deconvolution network for semantic segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1520–1528 (2015). https://doi.org/10.1109/ICCV.2015.178
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015). https://doi.org/10.1109/CVPR.2015.7298965
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495 (2017). https://doi.org/10.1109/TPAMI.2016.2644615
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017). https://doi.org/10.1145/3065386
Maas, A.L., Hannun, A.Y., Ng, A.Y.: Rectifier nonlinearities improve neural network acoustic models. In: Proceedings of ICML, p. 3 (2013)
Xu, B., Wang, N., Chen, T., Li, M.: Empirical evaluation of rectified activations in convolutional network. arXiv Preprint arXiv:1505.00853 (2015)
Gudi, S., et al.: Interobserver variability in the delineation of gross tumour volume and specified organs-at-risk during IMRT for head and neck cancers and the impact of FDG-PET/CT on such variability at the primary site. J. Med. Imaging Radiat. Sci. 48, 184–192 (2017). https://doi.org/10.1016/j.jmir.2016.11.003
Zhang, Q., Cui, Z., Niu, X., Geng, S., Qiao, Y.: Image segmentation with pyramid dilated convolution based on ResNet and U-Net. In: Liu, D., Xie, S., Li, Y., Zhao, D., El-Alfy, E.S. (eds.) Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), pp. 364–372. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-70096-0_38
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. 2818–2826. IEEE Computer Society (2016). https://doi.org/10.1109/CVPR.2016.308
Jégou, S., Drozdzal, M., Vazquez, D., Romero, A., Bengio, Y.: The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation (2017). https://openaccess.thecvf.com/content_cvpr_2017_workshops/w13/html/Jegou_The_One_Hundred_CVPR_2017_paper.html
Acknowledgements
M.A.N. is supported by a National Institutes of Health (NIH) Grant (R01 DE028290-01). K.A.W. is supported by a training fellowship from The University of Texas Health Science Center at Houston Center for Clinical and Translational Sciences TL1 Program (TL1 TR003169). C.D.F. received funding from the National Institute for Dental and Craniofacial Research Award (1R01DE025248-01/R56DE025248) and Academic-Industrial Partnership Award (R01 DE028290), the National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679), the NIH Big Data to Knowledge (BD2K) Program of the National Cancer Institute (NCI) Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01CA214825), the NCI Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01CA218148), the NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30CA016672), the NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007) and the National Institute of Biomedical Imaging and Bioengineering (NIBIB) Research Education Program (R25EB025787). He has received direct industry grant support, speaking honoraria and travel funding from Elekta AB.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Naser, M.A., van Dijk, L.V., He, R., Wahid, K.A., Fuller, C.D. (2021). Tumor Segmentation in Patients with Head and Neck Cancers Using Deep Learning Based-on Multi-modality PET/CT Images. In: Andrearczyk, V., Oreiller, V., Depeursinge, A. (eds) Head and Neck Tumor Segmentation. HECKTOR 2020. Lecture Notes in Computer Science(), vol 12603. Springer, Cham. https://doi.org/10.1007/978-3-030-67194-5_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-67194-5_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-67193-8
Online ISBN: 978-3-030-67194-5
eBook Packages: Computer ScienceComputer Science (R0)