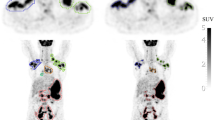
Abstract
Purpose
Total metabolic tumor volume (TMTV) segmentation has significant value enabling quantitative imaging biomarkers for lymphoma management. In this work, we tackle the challenging task of automated tumor delineation in lymphoma from PET/CT scans using a cascaded approach.
Methods
Our study included 1418 2-[18F]FDG PET/CT scans from four different centers. The dataset was divided into 900 scans for development/validation/testing phases and 518 for multi-center external testing. The former consisted of 450 lymphoma, lung cancer, and melanoma scans, along with 450 negative scans, while the latter consisted of lymphoma patients from different centers with diffuse large B cell, primary mediastinal large B cell, and classic Hodgkin lymphoma cases. Our approach involves resampling PET/CT images into different voxel sizes in the first step, followed by training multi-resolution 3D U-Nets on each resampled dataset using a fivefold cross-validation scheme. The models trained on different data splits were ensemble. After applying soft voting to the predicted masks, in the second step, we input the probability-averaged predictions, along with the input imaging data, into another 3D U-Net. Models were trained with semi-supervised loss. We additionally considered the effectiveness of using test time augmentation (TTA) to improve the segmentation performance after training. In addition to quantitative analysis including Dice score (DSC) and TMTV comparisons, the qualitative evaluation was also conducted by nuclear medicine physicians.
Results
Our cascaded soft-voting guided approach resulted in performance with an average DSC of 0.68 ± 0.12 for the internal test data from developmental dataset, and an average DSC of 0.66 ± 0.18 on the multi-site external data (n = 518), significantly outperforming (p < 0.001) state-of-the-art (SOTA) approaches including nnU-Net and SWIN UNETR. While TTA yielded enhanced performance gains for some of the comparator methods, its impact on our cascaded approach was found to be negligible (DSC: 0.66 ± 0.16). Our approach reliably quantified TMTV, with a correlation of 0.89 with the ground truth (p < 0.001). Furthermore, in terms of visual assessment, concordance between quantitative evaluations and clinician feedback was observed in the majority of cases. The average relative error (ARE) and the absolute error (AE) in TMTV prediction on external multi-centric dataset were ARE = 0.43 ± 0.54 and AE = 157.32 ± 378.12 (mL) for all the external test data (n = 518), and ARE = 0.30 ± 0.22 and AE = 82.05 ± 99.78 (mL) when the 10% outliers (n = 53) were excluded.
Conclusion
TMTV-Net demonstrates strong performance and generalizability in TMTV segmentation across multi-site external datasets, encompassing various lymphoma subtypes. A negligible reduction of 2% in overall performance during testing on external data highlights robust model generalizability across different centers and cancer types, likely attributable to its training with resampled inputs. Our model is publicly available, allowing easy multi-site evaluation and generalizability analysis on datasets from different institutions.








Similar content being viewed by others
Data Availability
The data that was used for model development (autoPET) is publicly available. To protect study participant privacy, we cannot share the testing data from BC Cancer, University of Wisconsin and Seoul St. Mary’s Hospital.
References
Cottereau A-S, Lanic H, Mareschal S, Meignan M, Vera P, Tilly H, et al. Molecular profile and FDG-PET/CT total metabolic tumor volume improve risk classification at diagnosis for patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2016;22:3801–9.
Kostakoglu L, Martelli M, Sehn LH, Belada D. Baseline PET-derived metabolic tumor volume metrics predict progression-free and overall survival in DLBCL after first-line treatment: results from the phase 3 …. Blood [Internet]. 2017; Available from: https://www.sciencedirect.com/science/article/pii/S000649711981340X.
Vercellino L, Cottereau A-S, Casasnovas O, Tilly H, Feugier P, Chartier L, et al. High total metabolic tumor volume at baseline predicts survival independent of response to therapy. Blood. 2020;135:1396–405.
Ceriani L, Martelli M, Zinzani PL, Ferreri AJM, Botto B, Stelitano C, et al. Utility of baseline 18FDG-PET/CT functional parameters in defining prognosis of primary mediastinal (thymic) large B-cell lymphoma. Blood. 2015;126:950–6.
Ceriani L, Milan L, Martelli M, Ferreri AJM, Cascione L, Zinzani PL, et al. Metabolic heterogeneity on baseline 18FDG-PET/CT scan is a predictor of outcome in primary mediastinal B-cell lymphoma. Blood. 2018;132:179–86.
Cottereau A-S, Versari A, Loft A, Casasnovas O, Bellei M, Ricci R, et al. Prognostic value of baseline metabolic tumor volume in early-stage Hodgkin lymphoma in the standard arm of the H10 trial. Blood. 2018;131:1456–63.
Mikhaeel NG, Smith D, Dunn JT, Phillips M, Møller H, Fields PA, et al. Combination of baseline metabolic tumour volume and early response on PET/CT improves progression-free survival prediction in DLBCL. Eur J Nucl Med Mol Imaging. 2016;43:1209–19.
Song M-K, Yang D-H, Lee G-W, Lim S-N, Shin S, Pak KJ, et al. High total metabolic tumor volume in PET/CT predicts worse prognosis in diffuse large B cell lymphoma patients with bone marrow involvement in rituximab era. Leuk Res. 2016;42:1–6.
Sasanelli M, Meignan M, Haioun C, Berriolo-Riedinger A, Casasnovas R-O, Biggi A, et al. Pretherapy metabolic tumour volume is an independent predictor of outcome in patients with diffuse large B-cell lymphoma. Eur J Nucl Med Mol Imaging. 2014;41:2017–22.
Toledano MN, Desbordes P, Banjar A, Gardin I, Vera P, Ruminy P, et al. Combination of baseline FDG PET/CT total metabolic tumour volume and gene expression profile have a robust predictive value in patients with diffuse large B-cell lymphoma. Eur J Nucl Med Mol Imaging. 2018;45:680–8.
Chang C-C, Cho S-F, Chuang Y-W, Lin C-Y, Chang S-M, Hsu W-L, et al. Prognostic significance of total metabolic tumor volume on 18F-fluorodeoxyglucose positron emission tomography/computed tomography in patients with diffuse large B-cell lymphoma receiving rituximab-containing chemotherapy. Oncotarget. 2017;8:99587–600.
Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, et al. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: the Lugano classification. J Clin Oncol. 2014;32:3059–68.
Ly J, Minarik D, Edenbrandt L, Wollmer P, Trägårdh E. The use of a proposed updated EARL harmonization of 18F-FDG PET-CT in patients with lymphoma yields significant differences in Deauville score compared with current EARL recommendations. EJNMMI Res. 2019;9:65.
Genc M, Yildirim N, Coskun N, Ozdemir E, Turkolmez S. The variation of quantitative parameters and Deauville scores with different reconstruction algorithms in FDG PET/CT imaging of lymphoma patients. Revista Española de Medicina Nuclear e Imagen Molecular (English Edition). 2023;42(6):388–92.
Ruppert AS, Dixon JG, Salles G, Wall A, Cunningham D, Poeschel V, et al. International prognostic indices in diffuse large B-cell lymphoma: a comparison of IPI, R-IPI, and NCCN-IPI. Blood. 2020;135:2041–8.
Meignan M, Cottereau A-S, Specht L, Mikhaeel NG. Total tumor burden in lymphoma — an evolving strong prognostic parameter. Br J Radiol. 2021;94:20210448.
El-Galaly TC, Villa D, Cheah CY, Gormsen LC. Pre-treatment total metabolic tumour volumes in lymphoma: does quantity matter? Br J Haematol. 2022;197:139–55.
Cottereau A-S, Meignan M, Nioche C, Capobianco N, Clerc J, Chartier L, et al. Risk stratification in diffuse large B-cell lymphoma using lesion dissemination and metabolic tumor burden calculated from baseline PET/CT†. Ann Oncol. 2021;32:404–11.
Alderuccio JP, Kuker RA, Barreto-Coelho P, Martinez BM, Miao F, Kwon D, et al. Prognostic value of presalvage metabolic tumor volume in patients with relapsed/refractory diffuse large B-cell lymphoma. Leuk Lymphoma. 2022;63:43–53.
Barrington SF, Meignan M. Time to prepare for risk adaptation in lymphoma by standardizing measurement of metabolic tumor burden. J Nucl Med. 2019;60:1096–102.
Hasani N, Paravastu SS, Farhadi F, Yousefirizi F, Morris MA, Rahmim A, et al. Artificial intelligence in lymphoma PET imaging: a scoping review (current trends and future directions). PET Clin. 2022;17:145–74.
Veziroglu EM, Farhadi F, Hasani N, Nikpanah M, Roschewski M, Summers RM, et al. Role of artificial intelligence in PET/CT imaging for management of lymphoma. Semin Nucl Med. 2023;53:426–48.
Burggraaff CN, Rahman F, Kaßner I, Pieplenbosch S, Barrington SF, Jauw YWS, et al. Optimizing workflows for fast and reliable metabolic tumor volume measurements in diffuse large B cell lymphoma. Mol Imaging Biol. 2020;22:1102–10.
Weisman AJ, Kieler MW, Perlman S, Hutchings M, Jeraj R, Kostakoglu L, et al. Comparison of 11 automated PET segmentation methods in lymphoma. Phys Med Biol. 2020;65:235019.
Huang L, Denœux T, Tonnelet D, Decazes P, Ruan S. Deep PET/CT fusion with Dempster-Shafer theory for lymphoma segmentation. Machine Learning in Medical Imaging. Springer International Publishing; 2021. p. 30–9.
Berthon B, Spezi E, Galavis P, Shepherd T, Apte A, Hatt M, et al. Toward a standard for the evaluation of PET — auto-segmentation methods following the recommendations of AAPM task group No. 211: Requirements and implementation. Med Phys. 2017;44:4098–111.
Ilyas H, Mikhaeel NG, Dunn JT, Rahman F, Møller H, Smith D, et al. Defining the optimal method for measuring baseline metabolic tumour volume in diffuse large B cell lymphoma. Eur J Nucl Med Mol Imaging. 2018;45:1142–54.
Hu H, Decazes P, Vera P, Li H, Ruan S. Detection and segmentation of lymphomas in 3D PET images via clustering with entropy-based optimization strategy. Int J Comput Assist Radiol Surg. 2019;14:1715–24.
Weisman AJ, Kim J, Lee I, McCarten KM, Kessel S, Schwartz CL, et al. Automated quantification of baseline imaging PET metrics on FDG PET/CT images of pediatric Hodgkin lymphoma patients. EJNMMI Phys. 2020;7:76.
Weisman AJ, Kieler MW, Perlman SB, Hutchings M, Jeraj R, Kostakoglu L, et al. Convolutional neural networks for automated PET/CT detection of diseased lymph node burden in patients with lymphoma. Radiol Artif Intell. 2020;2:e200016.
Blanc-Durand P, Jégou S, Kanoun S, Berriolo-Riedinger A, Bodet-Milin C, Kraeber-Bodéré F, et al. Fully automatic segmentation of diffuse large B cell lymphoma lesions on 3D FDG-PET/CT for total metabolic tumour volume prediction using a convolutional neural network. Eur J Nucl Med Mol Imaging. 2021;48:1362–70.
Shi T, Jiang H, Wang M, Diao Z, Zhang G, Yao YD. Metabolic anomaly appearance aware U-Net for automatic lymphoma segmentation in whole-body PET/CT scans. IEEE J Biomed Health Inform. 2023.
Yousefirizi F, Holloway C, Alexander A, Tonseth P, Uribe C, Rahmim A. Tumor segmentation of multi-centric whole-body PET/CT images from different cancers using a 3D convolutional neural network. J Nucl Med. 2022;63:2517–2517.
Jemaa S, Fredrickson J, Carano RAD, Nielsen T, de Crespigny A, Bengtsson T. Tumor segmentation and feature extraction from whole-body FDG-PET/CT using cascaded 2D and 3D convolutional neural networks. J Digit Imaging. 2020;33:888–94.
Hüllermeier E, Waegeman W. Aleatoric and epistemic uncertainty in machine learning: an introduction to concepts and methods. Mach Learn. 2021;110:457–506.
Gatidis S, Hepp T, Früh M, La Fougère C, Nikolaou K, Pfannenberg C, Schölkopf B, Küstner T, Cyran C, Rubin D. A whole-body FDG-PET/CT Dataset with manually annotated Tumor Lesions. Sci Data. 2022;9(1):601. Available from: https://wiki.cancerimagingarchive.net/x/LwKPBQ.
Zhang M, Levine S, Finn C. Memo: Test time robustness via adaptation and augmentation. Adv Neural Inf Process Syst. 2022;35:38629–42.
Matsunaga K, Hamada A, Minagawa A, Koga H. Image classification of melanoma, nevus and seborrheic keratosis by deep neural network ensemble. arXiv preprint arXiv:1703.03108. 2017;
Jin H, Li Z, Tong R, Lin L. A deep 3D residual CNN for false-positive reduction in pulmonary nodule detection. Med Phys. 2018;45:2097–107.
Jha AK, Bradshaw TJ, Buvat I, Hatt M, Prabhat KC, Liu C, Obuchowski NF, Saboury B, Slomka PJ, Sunderland JJ, Wahl RL. Nuclear medicine and artificial intelligence: best practices for evaluation (the RELAINCE guidelines). J Nucl Med. 2022;63(9):1288–99.
Saboury B, Bradshaw T, Boellaard R, Buvat I, Dutta J, Hatt M, et al. Artificial intelligence in nuclear medicine: opportunities, challenges, and responsibilities toward a trustworthy ecosystem. J Nucl Med. 2023;64:188–96.
Gatidis S, Früh M, Fabritius M, Gu S, Nikolaou K, La Fougère C, Ye J, He J, Peng Y, Bi L. The autoPET challenge: Towards fully automated lesion segmentation in oncologic PET/CT imaging. preprint at Research Square (Nature Portfolio). 2023.
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–57.
Gatidis S, Kuestner T. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions [Internet]. The Cancer Imaging Archive; 2022. Available from: https://wiki.cancerimagingarchive.net/x/LwKPBQ.
Shrestha A, Watkins A, Carlos U. RT-Utils: a minimal Python library to facilitate the creation and manipulation of DICOM RTStructs. GitHub; 2022. Available from: https://github.com/qurit/rt-utils/tree/main.
Zhang H, Cisse M, Dauphin YN, Lopez-Paz D. mixup: Beyond empirical risk minimization. arXiv preprint arXiv:1710.09412. 2017 Oct 25. cs.LG]. 2017. Available from: http://arxiv.org/abs/1710.09412.
Lee C-Y, Xie S, Gallagher P, Zhang Z, Tu Z. Deeply-supervised nets. In: Lebanon G, Vishwanathan SVN, editors. Proceedings of the Eighteenth International Conference on Artificial Intelligence and Statistics. San Diego: PMLR; 2015. p. 562–70.
Graziani M, Lompech T, Müller H, Depeursinge A, Andrearczyk V. On the scale invariance in state of the art CNNs trained on ImageNet. Mach Learn Knowl Extr. 2021;3:374–91.
Kim B, Ye JC. Mumford-Shah loss functional for image segmentation with deep learning. IEEE Trans Image Process. 2020;29:1856–66.
Yousefirizi F, Shiri I, Joo HO, Bloise I, Martineau P, Wilson D, et al. Semi-supervised learning towards automated segmentation of PET images with limited annotations: application to lymphoma patients [Internet]. arXiv [physics.med-ph]. 2022. Available from: http://arxiv.org/abs/2212.09908.
Yousefirizi F, Ahamed S, Joo HO, Bloise I, Saboury B, Rahmim A. Semi-supervised and unsupervised convolutional neural networks for automated lesion segmentation in PET imaging of lymphoma. J Nucl Med. 2022;63:3351.
Yousefirizi F, Dubljevic N, Ahamed S, Bloise I, Gowdy C, Joo HO, et al. Convolutional neural network with a hybrid loss function for fully automated segmentation of lymphoma lesions in FDG PET images. Medical Imaging 2022: Image Processing. SPIE; 2022. p. 214–20.
Huang L, Ruan S, Decazes P, Denœux T. Lymphoma segmentation from 3D PET-CT images using a deep evidential network. Int J Approx Reason. 2022;149:39–60.
Hatamizadeh A, Nath V, Tang Y, Yang D, Roth HR, Xu D. Swin UNETR: Swin transformers for semantic segmentation of brain tumors in MRI images. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing; 2022. p. 272–84.
Hadjiiski L, Cha K, Chan H-P, Drukker K, Morra L, Näppi JJ, et al. AAPM task group report 273: recommendations on best practices for AI and machine learning for computer-aided diagnosis in medical imaging. Med Phys. 2023;50:e1-24.
Bradshaw T, Boellaard R, Dutta J, Jha A, Jacobs P, Li Q, et al. Pitfalls in the development of artificial intelligence algorithms in nuclear medicine and how to avoid them. J Nucl Med. 2022;63:2724–2724.
Bradshaw TJ, Boellaard R, Dutta J, Jha AK, Jacobs P, Li Q, et al. Nuclear medicine and artificial intelligence: best practices for algorithm development. J Nucl Med [Internet]. 2021; Available from: https://doi.org/10.2967/jnumed.121.262567.
Yousefirizi F, Bloise I, Martineau P, Wilson D, Benard F, Bradshaw TB, et al. Reproducibility of a semiautomatic gradient-based segmentation approach for lymphoma PET. In: EANM abstract book, a supplement of the European Journal of Nuclear Medicine and Molecular Imaging (EJNMMI). Springer Science+Business Media; 2021.
Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–21.
Andrearczyk V, Oreiller V, Abobakr M, Akhavanallaf A, Balermpas P, Boughdad S, et al. Overview of the HECKTOR challenge at MICCAI 2022: automatic head and neck tumor segmentation and outcome prediction in PET/CT. Head Neck Tumor Chall. 2022;2023(13626):1–30.
Yousefirizi F, Jha AK, Brosch-Lenz J, Saboury B, Rahmim A. Toward high-throughput artificial intelligence-based segmentation in oncological PET imaging. PET Clin. 2021;16:577–96.
Funding
This research was in part supported by the Canadian Institutes of Health Research (CIHR) Project Grant PJT-173231, Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant RGPIN-2019–06467, GE Healthcare, and through computational resources and services provided by Microsoft for Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The retrospective study was conducted with the following ethics numbers: H19-01611 for the PMBCL study and H19-01866 for the DLBCL study and H19-001611 for Hodgkin study at BC Cancer, as well as KC11EISI0293 for the DLBCL study at Seoul St. Mary’s Hospital and UW2016-0418 for DLBCL and Hodgkin’s cases at University of Wisconsin.
Conflict of interest
Ivan Klyuzhin, Carlos Uribe, and Arman Rahmim are co-founders of Ascinta Technologies Inc.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yousefirizi, F., Klyuzhin, I.S., O, J.H. et al. TMTV-Net: fully automated total metabolic tumor volume segmentation in lymphoma PET/CT images — a multi-center generalizability analysis. Eur J Nucl Med Mol Imaging (2024). https://doi.org/10.1007/s00259-024-06616-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00259-024-06616-x