Abstract
Genetic studies of introduced non-native species are a valuable tool to investigate invasion history and pathways, source populations and multiple introductions of alien species, as well as evolutionary genetic changes following establishment in a new environment. We used a set of nine polymorphic microsatellites to analyse the population genetic structure of the introduced Asian shore crab Hemigrapsus takanoi along European coasts. Our dataset covered the complete known European range of the species, including the most recent records from Great Britain and the southwestern Baltic Sea. The results showed a similarly high genetic diversity of H. takanoi throughout Europe, and no indication of genetic bottlenecks during secondary spread, even in the most recently established populations. Analyses for population structure along geographic regions gave support for a separation between the Bay of Seine populations (northern France) and all other populations. Genetic differentiation within the North and Baltic Seas was more subtle and patchy, hinting to potential unrecognised introduction events, dispersal barriers and anthropogenic vector activity. The populations from the Baltic Sea and Great Britain clustered with the Wadden Sea populations, suggesting secondary introductions from the southeastern North Sea as likely invasion pathways. In summary, we suggest that a combination of anthropogenic secondary spread and the species’ reproductive biology have prevented a loss of genetic diversity during its ongoing expansion. We argue that genetic data depicting population status shortly after an introduction event—like the British and Baltic Sea populations of H. takanoi—may provide important baseline data for investigations of genetic changes during establishment and adaptation processes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Continuously rising numbers of human-mediated introductions of species into areas far beyond their native ranges are increasingly recognised as an important dimension of global change, especially in coastal and marine ecosystems (Ruiz et al. 2000; Chan and Briski 2017). Alien species are considered a major threat to native biodiversity and can profoundly alter invaded ecosystems (e.g., Bax et al. 2003; Molnar et al. 2008; Simberloff et al. 2013), warranting scientific attention towards new records of alien species and range expansions of established invaders.
Genetic markers are powerful tools to investigate biological invasions. They allow for the identification of source populations, invasion routes and the number of invasion events, as well as to understand dynamics of isolation, admixture and secondary spread in invaded ranges (Geller et al. 2010; Cristescu 2015). This is, in turn, valuable information for management strategies and invasion risk assessments (Kolar and Lodge 2001; Geller et al. 2010; Kulhanek et al. 2011). Furthermore, invasion genetics can provide insight into evolutionary processes following invasion and establishment in new ecosystems (Sakai et al. 2001; Lee 2002; Facon et al. 2008; Rius et al. 2015). In the latter context, genetic data from early invasion stages can provide important baselines for follow-up studies on longer time scales.
Multiple invasions, either from different areas in a species’ native range or repeatedly from the same source, as well as cryptic invasions, particularly the unrecognised introduction of novel genetic lineages, may play an important role for invasion success. Multiple introductions can alleviate detrimental effects of genetic drift and small founder populations on genetic diversity during invasion processes (Lockwood et al. 2005; Roman and Darling 2007; Geller et al. 2010; Lawson Handley et al. 2011). Admixture of genetically divergent lineages in invaded ranges can furthermore promote adaptation and accelerate range expansion (Rius and Darling 2014; Wagner et al. 2017). These phenomena appear to be rather common among marine invasive species. For example, the gastropod Cyclope neritea likely expanded along the French Atlantic coast as a result of multiple introductions from several regions within its native range since the 1970s (Simon-Bouhet et al. 2006). Similarly, Lejeusne et al. (2014) found no evidence of founder effects, but higher genetic diversity in invasive compared to native populations of the shrimp Palaemon macrodactylus, which they attributed to a combination of introductions from both native and already established invasive populations. The European shore crab Carcinus maenas, an infamous global invader, provides another prominent example: initially introduced to the NW Atlantic in the early 1800s, a rapid northward expansion and population growth along the Canadian Atlantic coast was observed since the 1980s, presumably as a consequence of multiple introductions from the northern native range (Roman 2006). The two distinct introduced lineages of C. maenas are now hybridising in the Canadian maritimes, potentially leading to increased thermal tolerance and overall increasing invasive potential (Jeffery et al. 2017; Lehnert et al. 2018). Genetic data also indicated recurring introductions of the Asian shore crab Hemigrapsus sanguineus in the NW Atlantic, and high genetic diversity of expanding invasive populations (Lord and Williams 2016; Blakeslee et al. 2017). The case of H. sanguineus further highlights the importance of intercontinental shipping as a vector for marine alien species (compare Gollasch 2006; Katsanevakis et al. 2013; Seebens et al. 2013), as intense ship traffic and the release of unexchanged ballast water from the native range coincided with the region of highest genetic diversity of H. sanguineus in the invaded range (Blakeslee et al. 2017).
Here, we explore the invasion genetics of Hemigrapsus takanoi, a recent invader of European coasts—in parts jointly with its congener H. sanguineus. Both Hemigrapsus spp. are highly successful marine invaders, even outnumbering and outcompeting C. maenas in its native and introduced range (Jensen et al. 2002; van den Brink et al. 2012; O’Connor 2014; Geburzi et al. 2018), as well as affecting the abundance of other benthic species (Kraemer et al. 2007; Brousseau et al. 2014). The native range of H. takanoi stretches along the coasts of the northwestern Pacific from southern Russia to China, including Japan and Taiwan (Asakura and Watanabe 2005; Yamasaki et al. 2011; Lee et al. 2013; Marin 2013). It inhabits the intertidal and shallow subtidal zones and also occurs in estuaries (Asakura and Watanabe 2005; Mingkid et al. 2006). Hemigrapsus takanoi was recorded in Europe for the first time in 1993 (then identified as H. penicillatus) in the hull fouling of a vessel docked at Bremerhaven in the Weser estuary/northern Germany (Gollasch 1999). While H. takanoi apparently failed to establish in the region at that time (Markert et al. 2014), a reproducing population was found at La Rochelle, France, in 1994 (Noël et al. 1997). By 1996, H. takanoi was recorded along the Bay of Biscay coast from northern Spain to the Loire estuary (Noël et al. 1997; Noël and Gruet 2008). In 1997, it was reported from Le Havre, northern France, and subsequently spread along the French coast of the English Channel (Breton et al. 2002; Dauvin 2009; Dauvin et al. 2009; Dauvin and Delhay 2010). In the North Sea, H. takanoi appeared for the first time in 2000 in the Dutch Delta (Nijland and Beekman 2000; Wolff 2005), followed by further records documenting a rapid spread along the Wadden Sea coast between 2006 (Texel/The Netherlands) and 2008 (Sylt/Germany) (Obert et al. 2007; Gittenberger et al. 2010; Landschoff et al. 2013). In 2014 and 2015, respectively, H. takanoi was reported from the southwestern Baltic Sea (Kiel Bight) and from southeastern Great Britain (Geburzi et al. 2015; Wood et al. 2015; Ashelby et al. 2017), and single individuals have been reported from the west coast of Sweden since 2017 (Karlsson et al. 2019), indicating an ongoing dynamic range expansion of this species. Markert et al. (2014) suggested three independent introductions of H. takanoi to Europe, to the Bay of Biscay, the French coast of the English Channel and the Dutch delta/southern North Sea. A recent study by Makino et al. (2018) found evidence for multiple introductions of this species, with the population in the Bay of Seine (northern France) being a genetic admixture of Chinese/Korean and Japanese origin, while the remaining European populations investigated were assigned exclusively to Japanese origin.
The recent invasion of the Baltic Sea by H. takanoi deserves particular attention, as this naturally species-poor, unique brackish water ecosystem is already strongly affected by alien species, which dominate benthic communities (Leppäkoski et al. 2002; Jazdzewski and Grabowski 2011; Ojaveer and Kotta 2015). For example, the crabs Eriocheir sinensis and Rhithropanopeus harrisii have strong ecological impacts (Rudnick and Resh 2005; Forsström et al. 2015). Considering the invasion potential and ecological impacts Hemigrapsus spp. has shown in other invaded regions (see above), the arrival of H. takanoi in the Baltic Sea warrants close monitoring.
We sampled H. takanoi populations throughout the species’ European range and used microsatellites for a regional-scale investigation of genetic structuring, with a special focus on shedding light on the pathways of the most recent expansions to Great Britain and the Baltic Sea, and on secondary dispersal within Europe in general. Based on the localities of initial introductions of H. takanoi suggested by Markert et al. (2014), we hypothesise that (i) populations from the North Sea, the French Bay of Biscay and English Channel coast will be genetically distinct, and (ii) the recently established populations in Great Britain and the Baltic Sea will be genetically similar to their geographically closest populations in the southwestern and southeastern North Sea, respectively. Within the Baltic Sea, we hypothesise (iii) to find genetic signatures of a stepwise expansion from the initial introduction site in Kiel Fjord (i.e., decreasing genetic diversity from west to east). Furthermore, we aim to gain insight in the mode and extent of secondary spread of H. takanoi within Europe (natural/larval dispersal vs. human-mediated spread) by investigating the geographic distribution of fine-scale genetic patterns.
Materials and methods
Sample collection and geographic information
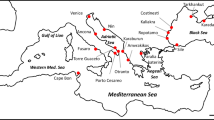
A total of 405 specimen of Hemigrapsus takanoi were collected from 13 sites along the coasts of the Wadden Sea (Germany/Netherlands), the southwestern Baltic Sea (Germany), the southwestern North Sea (United Kingdom) and the Bay of Biscay and Bay of Seine (France) between 2014 and 2018 (Table 1, Fig. 1). Sampling sites include the oldest known European population of H. takanoi in La Rochelle, France (site LRO), as well as the most recent reports from the Mecklenburg Bight in the German part of the Baltic Sea: Poel island near Wismar (site GOL, detected in autumn 2016, pers. comm. W. Wranik, University of Rostock), Neustadt/Holstein and the mouth of the Trave river near Lübeck (sites NEU and PRI, detected in summer 2017, this study). Crabs were hand collected between cobbles or mussels at low tide, or caught with a scoop from mussel clumps or underneath boulders in the shallow subtidal. All specimens were euthanised by rapid cooling (−80 °C for 15 min) and preserved as whole crabs in 96% ethanol. For better preservation of muscle tissue, the chelae and largest walking legs were perforated with a thin needle.
Expansion of H. takanoi in Europe and sampling sites used in this study. a Overview map with independent initial introductions (open circles) and expansion pathways (arrows) as hypothesised by Markert et al. (2014) and Geburzi et al. (2015, for the Baltic Sea). b–e Sampling sites by geographical region (see Table 1 for details and locality codes), with known years of first records. Colors for sampling sites correspond to Figs. 2, 3; colors are grouped by geographical region. The black star (d) indicates the location of the first ship-hull record in Europe (see “Introduction” for details)
DNA isolation, PCR amplification and microsatellite genotyping
Genomic DNA was extracted from pereiopod or chela muscle tissue using the Invisorb Spin Tissue Mini or Invisorb Spin Forensic kit (Stratec Molecular, Berlin, Germany). Eleven tri-nucleotide microsatellite loci (Ht02, Ht08, Ht12, Ht14, Ht20, Ht22, Ht28, Ht29, Ht34, Ht39, Ht47; Poux et al. 2015) were amplified in three multiplex sets by PCR in 15 µL volumes containing final concentrations of 0.9× PCR Buffer (Promega), 3.5 mM MgCl, 0.4 mM each dNTP (Roth), 0.3 µM each primer, 0.8 U GoTaq Polymerase (Promega) and 0.7–3 µL DNA template (depending on template concentration). PCR conditions were as follows: initial denaturation for 4 min at 95 °C, 30 cycles of denaturation for 30 s at 94 °C, annealing for 90 s at 57 °C, extension for 60 s at 72 °C, and a final extension of 20 min at 60 °C. PCR products were run on an AB 3730xl DNA Analyser (Applied Biosystems) with GeneScan 600 LIZ v2.0 size standard (Applied Biosystems) for fragment length analysis. Peaks were called and binned using the package Fragman 1.0.9 (Covarrubias-Pazaran et al. 2016) within R 3.4.2 (R Core Team 2017).
Analysis of genetic diversity and population structure
Genetic diversity in the samples was assessed by calculating the number of alleles per locus, observed and unbiased expected heterozygosity (HO and HE, Nei 1978), and allelic richness (Ar), the average number of alleles per population normalised to the smallest population size. Ar was calculated using the function ‘allel.rich’ of the R package PopGenReport 2.2.0 (Adamack and Gruber 2014). Deviations from Hardy–Weinberg equilibrium (HWE) were tested for each locus, both for each population separately and pooled over all populations, using the function ‘hw.test’ of the R package pegas 0.10 (Paradis 2010), with 1'000 Monte Carlo permutations. To correct for multiple comparisons, p values were corrected using the Benjamini–Hochberg method (Benjamini and Hochberg 1995). Frequency of null alleles for each locus was estimated based on the method of Brookfield (1996). Number of private alleles (i.e. alleles that occur only in one population) was calculated with the function ‘private_alleles’ implemented in the R package poppr 2.5.0 (Kamvar et al. 2014).
We assessed population structure by a multivariate discriminant analysis of principal components (DAPC, Jombart et al. 2010), implemented in the R package adegenet 2.1.2 (Jombart et al. 2008), and the Bayesian clustering algorithm STRUCTURE (Pritchard et al. 2000). The DAPC approach combines a principal component analysis (PCA) of the genotype data, which in a first step identifies alleles that best differentiate genotypes, and a discriminant analysis (DA), which in a second step uses the identified factors to maximise the variation between preassigned groupings. These groupings can be either defined by external criteria (i.e. biologically/geographically), or by clusters which are inferred directly from the data without any preliminary assumptions on population subdivision. The identification of the clusters is achieved by running k-means algorithm after PCA-transformation of the data. In adegenet, this is realised interactively by the function ‘find.clusters’. We conducted DAPCs following both approaches to assess if differences in the genotype data reflect geographic partitioning, and to detect possible genetic differentiation based on a priori unrecognised factors, respectively. Based on the retained discriminant functions, DAPC also calculates group membership probabilities for each individual. We used group membership probabilities from the DAPC approach with predefined groupings (i.e., sampling populations), to assess how clear-cut, or admixed, these populations genetically are.
Bayesian clustering was performed with STRUCTURE 2.3.4 (Pritchard et al. 2000; Falush et al. 2003, 2007; Hubisz et al. 2009), using the admixture ancestry model and the correlated allele frequency model. The sampling location was used as a prior in all analyses (‘LOCPRIOR’ function), as this improves the detection of clusters when the genetic structure is weak (Hubisz et al. 2009). We conducted the analysis hierarchically, examining the entire dataset (all 13 sampled populations) first, followed by two further levels of independent analyses of the clusters identified in the previous step. Each analysis was repeated 10 times for a number of clusters K, ranging from 1 to 1 + the number of populations in cluster, with 200′000 iterations after a burnin period of 20′000 iterations. We estimated the most likely numbers of clusters K based on the Evanno method (Evanno et al. 2005), as implemented in STRUCTURE HARVESTER 0.6.94 (Earl and VonHoldt 2012). Results of the replicated analyses were aligned using CLUMPP 1.1.2 (Jakobsson and Rosenberg 2007), and graphical output was produced using DISTRUCT 1.1 (Rosenberg 2004), both implemented in CLUMPAK (Kopelman et al. 2015).
To assess genetic differentiation between geographic and genetic groupings, we conducted an analysis of molecular variance (AMOVA, Excoffier et al. 1992), which is based on genetic distances between pairs of individual genotypes. Compared to the F-statistics measures of population differentiation (i.e., calculation of pairwise FST values), it has the advantage of not making assumptions about Hardy–Weinberg equilibrium and is thus also applicable for populations with significant homo- or heterozygote excess. For the H. takanoi microsatellite data, the genetic distance matrix was calculated based on relative dissimilarity (Prevosti et al. 1975), implemented in the R package poppr with the function ‘diss.dist’. We conducted hierarchical AMOVAs based on (1) geographical groupings (sampling population, region, sea; see Table 1), (2) clusters defined by k means algorithm without prior geographical information (see above), and (3) clusters defined by the hierarchical STRUCTURE analysis. The analyses were performed using the function ‘poppr.amova’ in the R package poppr. Significance of AMOVA results was assessed by randomising genotypes to populations in 1′000 Monte-Carlo permutations.
We estimated effective population size (Ne) from the genotype data as a fundamental parameter of population viability for each sampling population. For this, we used a bias-corrected linkage disequilibrium single-sample method (Waples 2006; Waples and Do 2008), as implemented in NeEstimator 2.1 (Do et al. 2014). We defined two critical values (Pcrit = 0.01 and 0.05) to filter out rare alleles, as they can strongly bias estimates of Ne (Do et al. 2014), and generated 95% confidence intervals using the jackknife across samples method (Jones et al. 2016) implemented in NeEstimator 2.1.
Results
Characterisation of the genotype data
We genotyped 326 out of the 405 initially collected individuals. As only very few (< 15) individuals were successfully scored on loci Ht28 and Ht47, these loci were removed from the dataset and not included in the analysis. All of the remaining nine loci were successfully scored on 240 individuals, more than 6 loci on 316 individuals, and more than 3 loci on 322 individuals. Scoring error rate was estimated to 4.28%, based on 37 individuals distributed over all sampling sites that were PCR amplified and scored a second time. The overall percentage of missing data was 4.99%, these results being consistent with the observed prevalence of null alleles (Table 2). Missing data were not evenly distributed over the dataset, but were especially prevalent in the loci Ht02 and Ht14 and populations LRO and TEX with > 8% missing data each. Thus, all analyses were performed on three datasets with different amounts of missing data (including individuals that amplified on more than three, more than six, and on all nine loci, respectively) and results were qualitatively the same. The results reported in the following are based on the analyses including those 322 individuals that scored on more than three loci (allele frequency table provided in Online Resource 1).
This dataset contained 7–32 individuals per population (Table 2). The nine microsatellite loci were highly polymorphic, total numbers of alleles per locus were 13–34. Allelic richness was similar in all populations, ranging between 5.92 and 7.53 (mean Ar per population over all loci). Mean expected heterozygosities varied between 0.81 and 0.89. Homozygote excess was prevalent, with significant deviations from HWE expectations on 3–8 loci in all populations except NEU and OUI. Estimated null allele frequencies ranged between 0.06 and 0.18. Conspicuously high numbers of private alleles were detected in the LRO (4), TEX (6) and PRI (8) populations (Table 2).
Genetic differentiation
DAPC
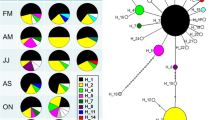
With the find.clusters function, three clusters were identified from the genotype data (BIC = 510.133 for k = 3, no. of retained PCs = 150). Results of the subsequent DAPC (no. of retained PCs = 75, no. of retained DAs = 2, proportion of conserved variance = 0.852), using the inferred clusters as group preassignments, showed no clear-cut pattern of cluster assignment in relation to the sampling populations (Fig. 2). Each cluster contained individuals from all populations, with some apparent patterning in the assignment proportions. While the Atlantic Ocean populations were predominantly assigned to cluster 1 (c. 60–75%), the northern and central Wadden Sea populations were predominantly assigned to cluster 2 (c. 41–65%). This pattern was not visible in the TEX and HAR populations, and the Baltic Sea populations showed mixed results (Table 3).
Discriminant analysis of principal components (DAPC) using clusters inferred from the genotype data as preassigned groupings, plot of the first two principal components. Point shape: cluster (1–3); point colour: sampling population of individual, corresponding to Fig. 1
The second DAPC approach, using the sampled populations as group preassignments, revealed some geographical structuring in the genetic data (no. of retained PCs = 95, no. of retained DAs = 12, proportion of conserved variance = 0.915). It clearly separated the Bay of Seine populations (HON and OUI) from all other populations. The remaining populations form one big cluster, with the central Wadden Sea populations (CUX, WHV, BEN) and the northern Wadden Sea, Southern Bight and Baltic Sea populations forming two very closely related sub-groups. The TEX population from the western Wadden Sea marginally separates from the cluster as a whole, but is well separated from the geographically closest central Wadden Sea populations (Fig. 3a). Only three alleles had a relative contribution greater than 0.05 to the observed clustering (Ht12:294, Ht22:165, Ht39:141; Fig. 3b), and their allele frequencies differed maximally about 0.35 between populations (Fig. 3c). The observed population structure is thus mainly the result of the frequency differences in these three alleles.
Discriminant analysis of principal components (DAPC) using sampling populations as preassigned groupings. a Plot of the first two axes of the DAPC; colour: population, corresponding to Fig. 1. b Bar graph of the relative contributions of alleles to the DACP principal components, alleles with a contribution > 0.05 (horizontal line) are indicated. c Line plot of allele frequencies with significant contributions for each population
The proportion of individuals successfully reassigned to their actual population based on the discriminant functions was 62.1% over the entire dataset, but varied strongly between populations. The recently recorded HAR population had the lowest reassignment proportion (20%) and showed high assignment probabilities to central and western Wadden Sea populations (Fig. 4a). In the recently invaded Baltic Sea, KIE and GOL (53.1% and 46.8% successful reassignments, respectively) appeared well admixed with each other and the Wadden Sea populations (Fig. 4b and c), but the remaining Baltic Sea populations NEU and PRI (57.1% and 76.0% successful reassignments, respectively) appeared as a more isolated, clear-cut group (Fig. 4b). The proportion of successful reassignments was overall highest in the Wadden Sea populations, varying between 53.1% (BEN) and 78.1% (LIS and TEX). These populations were clear-cut, and admixture appeared mostly with other Wadden Sea populations. The Bay of Seine populations OUI and HON showed intermediate proportions of successful reassignment (43.8% and 66.7%, respectively), but were admixed almost exclusively with each other, thus being clear-cut from all other populations. A complete overview of assignment probabilities for all populations is provided in Online Resource 2.
Assignment probabilities of individuals from the most recently recorded H. takanoi populations in Great Britain (a) and the Baltic Sea (b–d) as calculated by DAPC, when using sampling populations as groupings. Each row represents one individual, each column one group (i.e. sampling population). Colours represent assignment probability from 1 (red) to 0 (white), blue crosses indicate the original sampling population of the individual. The horizontal dashed line in c) separates the NEU and PRI individuals. See Table 1 for locality codes
Structure
The initial STRUCTURE analysis identified two major clusters (K = 2), separating OUI and HON (Bay of Seine) from all other populations (Fig. 5a). Qp values estimated by CLUMPP were > 85% for the Bay of Seine cluster, and > 90% for all populations in the other cluster, except for LRO (72%). No further substructure was detected in the Bay of Seine cluster, but a subsequent analysis of the remaining populations identified two clusters (K = 2), separating the central and northern Wadden Sea populations (LIS, CUX, WHV, BEN) from a cluster formed by the LRO, TEX, HAR and Baltic Sea populations. Subsequent analyses identified K = 3 clusters in the central and northern Wadden Sea group, matching well the sampling populations, and grouping together CUX and WHV (Fig. 5c). The cluster of CUX and WHV did not show any further substructure. For the group formed by the LRO, TEX, HAR and Baltic Sea populations, STRUCTURE identified K = 4 clusters, which showed LRO, TEX, and the group of NEU and PRI to be rather distinct (Qp > 70% for the dominating cluster), but highly admixed cluster memberships for HAR, KIE and GOL (Fig. 5c). No further substructure was detected among the latter three sampling populations.
Population structure as estimated from three levels of hierarchical STRUCTURE analysis. a Analysis over all sampling populations. b Separate analysis of the clusters formed by the Bay of Seine (right) and all other populations (left). c Separate analysis of the clusters formed by the northern and central Wadden Sea (left) and the LRO, TEX, HAR and Baltic Sea populations (right). Vertical coloured lines represent individual admixture coefficients; sampling populations are separated by vertical black lines
AMOVA
AMOVA results revealed only weak genetic differentiation on the basis of geographical groupings (i.e. sampling populations, regions or seas, Table 4 A). Variation between seas amounted to about 1.3%, variation between regions to about 0.8% (also being not significant), and variation between populations to about 2.1% of total variation in the genotype data. However, with clusters identified by k-means as grouping, AMOVA revealed that 5.7% of variation is explained by differences between these clusters (Table 4 B). Likewise, the AMOVA based on groupings by the initial STRUCTURE analysis (K = 2) explained 4.79% of the total variation by differences between the two clusters, and another 2.61% by differences between populations within these clusters. This indicates that, on average, the Bay of Seine populations (OUI and HON) differ genetically from the North and Baltic Sea populations by about 7.4% (Table 4C). Genetic differentiation among the clusters identified in the subsequent STRUCTURE analyses was much lower (max. 1.09%), but still significant. In all separate analyses, the highest proportion of variation was detected within populations, explaining between 92.3% and 97.8% of total genetic variation (Table 4).
Effective population size
Calculations of Ne by linkage disequilibrium method (LDNe) with a random mating model estimated infinite Ne values for six and eight of the sampling populations, for lowest allele frequencies of 0.05 and 0.01, respectively. In particular, no reduced Ne was detected for the recently established populations from Great Britain (HAR) and the Baltic Sea (KIE, NEU, PRI and GOL). The upper 95% confidence interval was estimated to be infinite by the jackknife on samples method for all populations. This was also the case for the HON population with a low Ne of 37.0 and 29.6 for lowest allele frequencies of 0.05 and 0.01, respectively (Table 5).
Discussion
We found similarly high levels of genetic diversity (measured as allelic richness) in all sampled European populations of H. takanoi. Although not directly comparable due to the different microsatellites used, these values are in ranges of those reported for invasive H. sanguineus populations in the NW Atlantic (Blakeslee et al. 2017). Invasive populations rather commonly maintain high levels of genetic diversity during the primary invasion process, putatively as a consequence of large, genetically diverse founding populations or recurring migration/introduction to the newly colonised regions (reviewed in Roman and Darling 2007; Lawson Handley et al. 2011). While this could be assumed to apply to H. takanoi as well, considering the high potential for ballast water and hull fouling transport of large numbers of propagules from Asia to Europe (compare Gollasch 1999; Seebens et al. 2013; Blakeslee et al. 2017), Makino et al. (2018) found in fact evidence for a genetic bottleneck during the primary introduction of H. takanoi. Secondary spread of marine invasive species within the invaded range however regularly leads to a loss of genetic diversity as a consequence of consecutive bottlenecks (e.g. Herborg et al. 2007; Tepolt et al. 2009; Reusch et al. 2010; Pérez-Portela et al. 2012). Notably, we did not find genetic diversity to be reduced in the most recently detected Baltic Sea and Great Britain populations of H. takanoi. A lack of secondary bottleneck effects was also observed in H. sanguineus during its northward expansion along the US east coast (Lord and Williams 2016), as well as in R. harrisii expanding into the northern Baltic Sea (Forsström et al. 2017). Interestingly, females of all three crab species are capable of sperm storage (Anderson and Epifanio 2010; Rodgers et al. 2011; van den Brink et al. 2013), and multiple mating can occur in both Hemigrapsus spp. (Anderson and Epifanio 2010, own observations). A single ovigerous female introduced to a new region, as we for example observed in Kiel Fjord in 2014 (Geburzi et al. 2015), may, therefore, introduce a large number of genetically diverse propagules. This provides a potential mechanism for Hemigrapsus spp. and other crab species with a similar reproductive strategy to avoid bottlenecks during secondary spread despite small founding populations (compare Eales et al. 2010).
Our data indicate only weak overall population differentiation of European H. takanoi for groupings defined by either geography or genetic clustering methods, with by far the highest share of genetic variation occurring within sampled populations. Such a pattern of high genetic diversity and weak population structure was also observed in several other systems, and has been attributed to multiple introductions from divergent sources (Simon-Bouhet et al. 2006), high dispersal ability by planktonic larvae (Viard et al. 2006), and a rapid multidirectional expansion after a very recent introduction (compare Fitzpatrick et al. 2012; Gonçalves et al. 2019). In fact, more pronounced genetic structure in invaded ranges seems to occur more regularly in species with a longer invasion history (i.e. several decades), e.g., E. sinensis (Hänfling et al. 2002; Hayer et al. 2019) and R. harrisii (Forsström et al. 2017) in Europe. Future genetic studies of H. takanoi, comparing longer time spans, could be used to show if genetic structure establishes—or is prevented by recurring introductions or natural/anthropogenic mixing.
One factor potentially obscuring our results is the prevalence of null alleles on all loci (estimated frequency 0.06–0.18). Null alleles are caused by mutations in the microsatellite flanking regions, hampering the binding of primers during PCR amplification, thus leading to loci falsely scored as homozygous. Accordingly, we detected the strongest deviations from Hardy–Weinberg expectations on loci with the highest frequency of null alleles, providing a likely explanation for the observed homozygote excess. Null alleles are also a likely cause for the almost complete failure to amplify two loci (Ht28 and Ht47) from the initial set of microsatellites. Chapuis (2007) showed that the frequency of null alleles increases with increasing effective population size, as the probability for mutations in microsatellite flanking regions rises. This is in accordance with our large estimates of Ne for the majority of sampled populations. They, in turn, can be explained by the species’ reproductive biology, particularly its early maturity and the possibility of interbreeding between subsequent cohorts (Gothland et al. 2014), which likely contribute to the rapid population growth and high densities observed in invasive H. takanoi populations (van den Brink et al. 2012; Gothland et al. 2014; Geburzi et al. 2018).
The most distinct feature of population structure detected by our analyses is the clear separation of the Bay of Seine (sampling populations OUI and HON) from all other regions included in this study. This separation was recovered by both, DAPC and STRUCTURE analyses, and the assignment tests indicated only very little admixture between the Bay of Seine and the other regions. The distinctness of the Bay of Seine could point to an independent introduction from the native range into this region, and is in fact in accordance with the results of Makino et al. (2018) who genetically compared native and invasive populations of H. takanoi. It is curious that this pattern seems to be temporally stable (the specimens for their study were collected between 2012 and 2014, ours in 2016), despite the fact that the Bay of Seine population is likely formed by hybrids from two genetically distinct sources in the native range (Makino et al. 2018). Such admixture events have the potential to promote rapid expansion of invasive populations, e.g., by heterosis effects that enhance adaptation to novel environmental conditions (Kelly et al. 2006; Rius and Darling 2014; Wagner et al. 2017). Larval dispersal of H. takanoi from the Bay of Seine might be restricted by the local current system that forms a gyre at the western entrance of the bay (Salomon and Breton 1993). Lefebvre et al. (2003) showed that these conditions lead to strong retention of larvae of the brittle star Ophiotrix fragilis in the Bay of Seine, and this mechanism may apply to H. takanoi larvae as well. However, the large port of Le Havre is directly connected to various ports along the British and continental European North Sea coast (data from www.marinetraffic.org, accessed daily between 2020-02-17 and 2020-02-23) via shipping activities, and anthropogenic secondary spread of H. takanoi from this region may occur in the future.
In contrast to our initial hypothesis (i), we did not find H. takanoi from the French Bay of Biscay coast (LRO) to be genetically distinct from H. takanoi from the North Sea. This does not necessarily imply the invasion of H. takanoi into both regions to be the result of the same introduction event, as Gollasch (1999) speculated—particularly given the 6 years time difference between the first reports of established H. takanoi populations in the Bay of Biscay in 1994 (Noël et al. 1997) and the Dutch Delta in 2000 (Nijland and Beekman 2000). Independent introductions from the same region in the native range (being in accordance with the hypothesis of Markert et al. 2014), or secondary spread from the Bay of Biscay to the North Sea are possible explanations for their genetic similarity, but this question remains unresolved by our data.
For the North Sea/Baltic Sea system, our results show a high genetic relatedness of the recently established populations in Great Britain (HAR) and in the Baltic Sea with the Wadden Sea populations, which is in accordance to our hypothesis (ii). This indicates secondary spread from the southeastern North Sea to be the most likely introduction pathway of H. takanoi to both regions. Individuals from the HAR population showed increased assignment probabilities to sites in the western and central Wadden Sea, respectively, hinting to possible admixture from these two regions at the British coast. This could promote further expansion of H. takanoi in this region by increasing the adaptive potential/environmental tolerance of the admixed population (compare Kelly et al. 2006; Wagner et al. 2017). However, we are cautious with this conclusion, as our data indicate only little overall divergence between the western and central Wadden Sea. The introduction to Great Britain and the Baltic Sea has likely been mediated by anthropogenic transport rather than larval dispersal. Residual coastal currents in the southern North Sea run southwest to northeast from the French Channel along the Dutch Delta, Wadden Sea and Danish coasts, while they run north to south along the British coast (Wolff 2005; Winther and Johannessen 2006), rendering current-mediated larval spread from east to west across the southern North Sea rather unlikely. Likewise, H. takanoi reached the Baltic Sea most likely by transportation through the Kiel Canal rather than by larval dispersal around the Danish peninsula, considering the lack of records from the coasts of Denmark, as well as the fact that H. takanoi was found in Kiel Fjord (at the Baltic Sea entrance of the canal) three years earlier than on the west coast of Sweden (Geburzi et al. 2015; Karlsson et al. 2019). These assumptions are supported by the high genetic diversity we found in these very young populations, indicating larger founding populations (see above).
Interestingly, our DAPC and STRUCTURE analysis showed some genetic differentiation between H. takanoi in the western Wadden Sea (TEX) and the central/northern Wadden Sea group (BEN, WHV, CUX and LIS), despite being geographically close to each other. The planktonic larval duration of H. takanoi is at least 22 days (at 20 °C water temperature, Landeira et al. 2019). This should enable long-distance dispersal along the Wadden Sea coast mediated by the current system described above, which would be expected to counteract genetic differentiation within the Wadden Sea. Similar to our results, Moehler et al. (2011) found populations of the invasive Pacific oyster Magallana gigas in the northern Wadden Sea to be genetically distinct from those in other parts of the Wadden Sea, which they attributed to different source populations for oyster stockings. In the case of H. takanoi, the observed pattern might also hint to separate introductions to the North Sea: One introduction site could have been the Dutch Delta, from where H. takanoi spread to the western Wadden Sea (compare Markert et al. 2014), while another introduction could have occurred to a port along the central Wadden Sea coast (e.g., Hamburg, Bremerhaven, compare Gollasch 1999). Differential local adaptation could then have occurred in different parts of the North Sea, “blocking” the establishment of propagules from the western Wadden Sea/Dutch Delta region in the central and northern Wadden Sea by intraspecific priority effects (sensu Makino et al. 2018). The possibility of contrasting invasion trajectories within the Wadden Sea ecosystem was for example demonstrated for the parasitic copepod Mytilicola intestinalis (Feis et al. 2016). Alternatively, the outflow of the Ems river might form a physical barrier for H. takanoi larval dispersal from the western to the central Wadden Sea. More detailed, fine scale studies, including the Dutch Delta as potential introduction site, would be needed to answer these questions.
Within the Baltic Sea, we hypothesised (iii) a decline of genetic diversity from west to east, as a result of consecutive bottlenecks following the rapid dispersal from the initial introduction site (KIE, detected in 2014) to Mecklenburg Bight (GOL, detected in 2016). However, our data showed no reduction of genetic diversity in any of the Baltic Sea sampling populations, leading us to conclude that anthropogenic dispersal contributed to the secondary spread of H. takanoi in the Baltic Sea. Circumstantial evidence supports this conclusion: first, residual surface currents in the southwestern Baltic Sea predominantly run east to west/northwest (Karlsson et al. 2019), a pattern that is only occasionally reverted by long-lasting, strong westerly winds (Svansson 1975). Second, a snapshot survey along the coasts of Kiel and Mecklenburg Bight in 2017 revealed a currently patchy distribution of H. takanoi in the southwestern Baltic Sea, with gaps between occupied sites despite the availability of suitable habitat (Weiler and Geburzi, unpublished data). We would expect a more continuous distribution along the coastline if the spread was mediated by natural processes alone. Also, the fact that H. takanoi occurs almost exclusively in marinas or their close vicinity hints to a human-mediated spread within the Baltic Sea. The potential of recreational boating acting as vector for the secondary spread of invasive species was demonstrated by a number of studies, including the transfer of mobile crustacean species in fouling communities and residual water (e.g., Darbyson et al. 2009; Clarke Murray et al. 2011; Kelly et al. 2013). Furthermore, the sampled populations in Lübeck Bight (PRI and NEU) appeared curiously distinct from the other sites in the Baltic Sea, as indicated by our STRUCTURE analysis, assignment tests, as well as the high prevalence of private alleles. This finding might hint to an independent introduction to this region, rather than a stepwise spread of H. takanoi from Kiel Fjord to Lübeck Bight and further east. Overall, the genetic pattern observed in the Baltic Sea (and in the European range of H. takanoi in general), resembles that of a “stratified dispersal”, consisting of a combination of short-range natural dispersal, and occasional anthropogenic dispersal “jumps” (Bronnenhuber et al. 2011).
Conclusions and directions for future research
Within 25 years since its first records in Europe, Hemigrapsus takanoi has expanded successfully along several thousand kilometres of European Atlantic, North Sea and—most recently—Baltic Sea coast line. Throughout this expansion process, H. takanoi apparently did not suffer losses in genetic diversity, which we interpret as an important contribution to the species’ invasion success, as maintaining high levels of genetic diversity during invasions can enhance the invaders’ ability to adapt to novel selective pressures (Lee 2002; Welles and Dlugosch 2018). We found that characteristics of the reproductive biology of H. takanoi and circumstantial evidence for anthropogenic secondary spread within Europe are likely explanations for the overall patterns of high genetic diversity and weak genetic differentiation we observed in our data. The interplay of these factors might be a common feature shared with other invasive crabs, contributing to the high proportion of successful invasive species in the Brachyura (Brockerhoff and McLay 2011).
In the Baltic Sea in particular, the high genetic diversity detected in the very recent invader H. takanoi contrasts to a common pattern of many native Baltic Sea species. These often show a reduced genetic diversity and high genetic isolation compared to their conspecific (and putative ancestral) populations in the North Atlantic, as a consequence of bottlenecks and adaptation/selection (Johannesson and André 2006). Our results indicate that human-mediated introductions might override this natural barrier for colonisation of the Baltic Sea and could contribute to the overall high number of successful invasions in the Baltic Sea. We, therefore, assume a high expansion potential of H. takanoi in the Baltic Sea. In this context, the population in the Kattegat at the west coast of Sweden should be included in future genetic studies, and the Danish archipelago should be closely monitored, given the potential of admixture between expanding Kattegat and southwestern Baltic populations in this region.
Overall, our study contributes to the detection of common marine invasion trajectories in European Seas to inform management strategies. It furthermore showcases the potential of invasive species to maintain genetic diversity during rapid secondary expansion, a phenomenon that might well occur in other species with similar reproductive traits and/or areas with similarly high regional vector activity. Our study thus highlights the power of combining invasion genetics with studies on an invader’s general biology and dispersal pathways to fully understand successful invasion processes and their outcomes. However, it also showed some limitations of microsatellite markers to fully resolve very recent invasion and expansion dynamics. The application of genomic markers could overcome these limitations in future studies, and would also allow to assess non-neutral evolutionary outcomes of the species’ invasion. The results of this study furthermore provide data on the genetic diversity and structure of European H. takanoi populations from a very early invasion stage, which may form a unique, well-documented baseline for long-term investigations of genetic changes during establishment and adaptation processes.
Data availability
The complete microsatellite allele frequency data are provided in the supplementary electronic material (online resource 1) of this publication.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387. https://doi.org/10.1111/2041-210X.12158
Anderson JA, Epifanio CE (2010) Mating and sperm storage of the Asian shore crab Hemigrapsus sanguineus. J Shellfish Res 29:497–501. https://doi.org/10.2983/035.029.0228
Asakura A, Watanabe S (2005) Hemigrapsus takanoi, new species, a sibling species of the common Japanese intertidal crab H. penicillatus (Decapoda: Brachyura: Grapsoidea). J Crustac Biol 25:279–292. https://doi.org/10.1651/C-2514
Ashelby CW, Sewell J, Rostron J, Shrubsole R, Child T, Clark PF (2017) Evidence for the invasion and successful establishment of Hemigrapsus takanoi Asakura and Watanabe, 2005 (Decapoda, Varunidae) in Great Britain. Crustaceana 90:695–708. https://doi.org/10.1163/15685403-00003692
Bax NJ, Williamson A, Aguero M, Gonzalez E, Geeves W (2003) Marine invasive alien species: a threat to global biodiversity. Mar Policy 27:313–323
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. https://doi.org/10.2307/2346101
Blakeslee AMH, Kamakura Y, Onufrey J, Makino W, Urabe J, Park S, Keogh CL, Whitman Miller A, Minton MS, Carlton JT, Miura O (2017) Reconstructing the Invasion History of the Asian shorecrab, Hemigrapsus sanguineus (De Haan 1835) in the Western Atlantic. Mar Biol 164:47. https://doi.org/10.1007/s00227-017-3117-x
Breton G, Faasse M, Noël PY, Vincent T (2002) A new alien crab in Europe: Hemigrapsus sanguineus (Decapoda: Brachyura: Grapsidae). J Crustac Biol 22:184–189. https://doi.org/10.1651/0278-0372(2002)022[0184:ANACIE]2.0.CO;2
Brockerhoff AM, McLay CL (2011) Human-Mediated Spread of Alien Crabs. In: Galil BS, Clark PF, Carlton JT (eds) In the Wrong Place - Alien Marine Crustaceans: distribution, Biology and Impacts. Springer, Netherlands, pp 27–106
Bronnenhuber JE, Dufour BA, Higgs DM, Heath DD (2011) Dispersal strategies, secondary range expansion and invasion genetics of the nonindigenous round goby, Neogobius melanostomus, in Great Lakes tributaries. Mol Ecol 20:1845–1859. https://doi.org/10.1111/j.1365-294X.2011.05030.x
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–455. https://doi.org/10.1046/j.1365-294X.1996.00098.x
Brousseau DJ, Goldberg R, Garza C (2014) Impact of Predation by the Invasive Crab Hemigrapsus sanguineus on Survival of Juvenile Blue Mussels in Western Long Island Sound. Northeast Nat 21:119–133. https://doi.org/10.1656/045.021.0110
Chan FT, Briski E (2017) An overview of recent research in marine biological invasions. Mar Biol 164:121. https://doi.org/10.1007/s00227-017-3155-4
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631. https://doi.org/10.1093/molbev/msl191
Clarke Murray C, Pakhomov EA, Therriault TW (2011) Recreational boating: a large unregulated vector transporting marine invasive species. Divers Distrib 17:1161–1172. https://doi.org/10.1111/j.1472-4642.2011.00798.x
Covarrubias-Pazaran G, Diaz-Garcia L, Schlautman B, Salazar W, Zalapa J (2016) Fragman: an R package for fragment analysis. BMC Genet 17:62. https://doi.org/10.1186/s12863-016-0365-6
Cristescu ME (2015) Genetic reconstructions of invasion history. Mol Ecol 24:2212–2225. https://doi.org/10.1111/mec.13117
Darbyson E, Locke A, Hanson JM, Willison JHM (2009) Marine boating habits and the potential for spread of invasive species in the Gulf of St. Lawrence Aquat Invasions 4:87–94. https://doi.org/10.3391/ai.2009.4.1.9
Dauvin J-C (2009) Asian Shore Crabs Hemigrapsus spp. (Crustacea: Brachyura: Grapsoidea) continue their invasion around the Cotentin Peninsula, Normandy, France: status of the Hemigrapsus population in 2009. Aquat Invasions 4:605–611. https://doi.org/10.3391/ai.2009.4.4.6
Dauvin J-C, Delhay J-B (2010) First record of Hemigrapsus takanoi (Crustacea: Decapoda: Grapsidae) on the western coast of northern Cotentin, Normandy, western English Channel. Mar Biodivers Rec 3:1–3. https://doi.org/10.1017/S1755267210000928
Dauvin J-C, Tous Rius A, Ruellet T (2009) Recent expansion of two invasive crabs species Hemigrapsus sanguineus (de Haan, 1835) and H. takanoi Asakura and Watanabe 2005 along the Opal Coast. France Aquat Invasions 4:451–465. https://doi.org/10.3391/ai.2009.4.3.3
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214. https://doi.org/10.1111/1755-0998.12157
Eales J, Thorpe RS, Malhotra A (2010) Colonization history and genetic diversity: adaptive potential in early stage invasions. Mol Ecol 19:2858–2869. https://doi.org/10.1111/j.1365-294X.2010.04710.x
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131:479–491. https://doi.org/10.1007/s00424-009-0730-7
Facon B, Pointier JP, Jarne P, Sarda V, David P (2008) High genetic variance in life-history strategies within invasive populations by way of multiple introductions. Curr Biol 18:363–367. https://doi.org/10.1016/j.cub.2008.01.063
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578. https://doi.org/10.1111/j.1471-8286.2007.01758.x
Feis ME, Goedknegt MA, Thieltges DW, Buschbaum C, Wegner KM (2016) Biological invasions and host-parasite coevolution: different coevolutionary trajectories along separate parasite invasion fronts. Zoology 119:366–374. https://doi.org/10.1016/j.zool.2016.05.012
Fitzpatrick BM, Fordyce JA, Niemiller ML, Reynolds RG (2012) What can DNA tell us about biological invasions? Biol Invasions 14:245–253. https://doi.org/10.1007/s10530-011-0064-1
Forsström T, Fowler AE, Manninen I, Vesakoski O (2015) An introduced species meets the local fauna: predatory behavior of the crab Rhithropanopeus harrisii in the Northern Baltic Sea. Biol Invasions 17:2729–2741. https://doi.org/10.1007/s10530-015-0909-0
Forsström T, Ahmad F, Vasemägi A (2017) Invasion genomics: genotyping-by-sequencing approach reveals regional genetic structure and signatures of temporal selection in an introduced mud crab. Mar Biol 164:186. https://doi.org/10.1007/s00227-017-3210-1
Geburzi JC, Graumann G, Köhnk S, Brandis D (2015) First record of the Asian crab Hemigrapsus takanoi Asakura and Watanabe, 2005 (Decapoda, Brachyura, Varunidae) in the Baltic Sea. BioInvasions Rec 4:103–107. https://doi.org/10.3391/bir.2015.4.2.06
Geburzi JC, Brandis D, Buschbaum C (2018) Recruitment patterns, low cannibalism and reduced interspecific predation contribute to high invasion success of two Pacific crabs in northwestern Europe. Estuar Coast Shelf Sci 200:460–472. https://doi.org/10.1016/j.ecss.2017.11.032
Geller JB, Darling JA, Carlton JT (2010) Genetic perspectives on marine biological invasions. Ann Rev Mar Sci 2:367–393. https://doi.org/10.1146/annurev.marine.010908.163745
Gittenberger A, Rensing M, Stegenga H, Hoeksema B (2010) Native and non-native species of hard substrata in the Dutch Wadden Sea. Ned Faun Meded 33:21–76
Gollasch S (1999) The Asian decapod Hemigrapsus penicillatus (De Haan, 1835) (Grapsidae, Decapoda) introduced in European waters: status quo and future perspective. Helgoländer Meeresuntersuchungen 52:359–366. https://doi.org/10.1007/BF02908909
Gollasch S (2006) Overview on introduced aquatic species in European navigational and adjacent waters. Helgol Mar Res 60:84–89. https://doi.org/10.1007/s10152-006-0022-y
Gonçalves RM, Mastrangelo T, Rodrigues JCV, Paulo DF, Omoto C, Corrêa AS, de Azeredo-Espin AML (2019) Invasion origin, rapid population expansion, and the lack of genetic structure of cotton bollworm (Helicoverpa armigera) in the Americas. Ecol Evol 9:7378–7401. https://doi.org/10.1002/ece3.5123
Gothland M, Dauvin J-C, Denis L, Dufossé F, Jobert S, Ovaert J, Pezy J-P, Tous Rius A, Spilmont N (2014) Biological traits explain the distribution and colonisation ability of the invasive shore crab Hemigrapsus takanoi. Estuar Coast Shelf Sci 142:41–49. https://doi.org/10.1016/j.ecss.2014.03.012
Hänfling B, Carvalho GR, Brandl R (2002) mt-DNA sequences and possible invasion pathways of the Chinese mitten crab. Mar Ecol Prog Ser 238:307–310
Hayer S, Brandis D, Hartl GB, Ewers-Saucedo C (2019) First indication of Japanese mitten crabs in Europe and cryptic genetic diversity of invasive Chinese mitten crabs. NeoBiota 50:1–29. https://doi.org/10.3897/neobiota.50.34881
Herborg L-M, Weetman D, van Oosterhout C, Hänfling B (2007) Genetic population structure and contemporary dispersal patterns of a recent European invader, the Chinese mitten crab, Eriocheir sinensis. Mol Ecol 16:231–242. https://doi.org/10.1111/j.1365-294X.2006.03133.x
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332. https://doi.org/10.1111/j.1755-0998.2009.02591.x
Jakobsson M, Rosenberg NA (2007) CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. https://doi.org/10.1093/bioinformatics/btm233
Jazdzewski K, Grabowski M (2011) Alien Crustaceans Along the Southern and Western Baltic Sea. In: Galil BS, Clark PF, Carlton JT (eds) In the Wrong Place - Alien Marine Crustaceans: Distribution, Biology and Impacts. Springer, Netherlands, Dordrecht, pp 323–344
Jeffery NW, DiBacco C, Wringe BF, Stanley RRE, Hamilton LC, Ravindran PN, Bradbury IR (2017) Genomic evidence of hybridization between two independent invasions of European green crab (Carcinus maenas) in the Northwest Atlantic. Heredity (Edinb) 119:154–165. https://doi.org/10.1038/hdy.2017.22
Jensen GC, McDonald PS, Armstrong DA (2002) East meets west: competitive interactions between green crab Carcinus maenas, and native and introduced shore crab Hemigrapsus spp. Mar Ecol Prog Ser 225:251–262. https://doi.org/10.3354/meps225251
Johannesson K, André C (2006) Life on the margin: Genetic isolation and diversity loss in a peripheral marine ecosystem, the Baltic Sea. Mol Ecol 15:2013–2029. https://doi.org/10.1111/j.1365-294X.2006.02919.x
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Jones AT, Ovenden JR, Wang YG (2016) Improved confidence intervals for the linkage disequilibrium method for estimating effective population size. Heredity (Edinb) 117:217–223. https://doi.org/10.1038/hdy.2016.19
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Karlsson R, Obst M, Berggren M (2019) Analysis of potential distribution and impacts for two species of alien crabs in Northern Europe. Biol Invasions 21:3109–3119. https://doi.org/10.1007/s10530-019-02044-3
Katsanevakis S, Zenetos A, Belchior C, Cardoso AC (2013) Invading European Seas: Assessing pathways of introduction of marine aliens. Ocean Coast Manag 76:64–74. https://doi.org/10.1016/j.ocecoaman.2013.02.024
Kelly DW, Muirhead JR, Heath DD, Macisaac HJ (2006) Contrasting patterns in genetic diversity following multiple invasions of fresh and brackish waters. Mol Ecol 15:3641–3653. https://doi.org/10.1111/j.1365-294X.2006.03012.x
Kelly NE, Wantola K, Weisz E, Yan ND (2013) Recreational boats as a vector of secondary spread for aquatic invasive species and native crustacean zooplankton. Biol Invasions 15:509–515. https://doi.org/10.1007/s10530-012-0303-0
Kolar CS, Lodge DM (2001) Progress in invasion biology: predicting invaders. Trends Ecol Evol 16:199–204
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Kraemer GP, Sellberg M, Gordon A, Main J (2007) Eight-year Record of Hemigrapsus sanguineus (Asian Shore Crab) Invasion in Western Long Island Sound Estuary. Northeast Nat 14:207–224. https://doi.org/10.1656/1092-6194(2007)14[207:EROHSA]2.0.CO;2
Kulhanek SA, Ricciardi A, Leung B (2011) Is invasion history a useful tool for predicting the impacts of the world’s worst aquatic invasive species? Ecol Appl 21:189–202. https://doi.org/10.2307/29779646
Landeira JM, Cuesta JA, Tanaka Y (2019) Larval development of the brush-clawed shore crab Hemigrapsus takanoi Asakura & Watanabe, 2005 (Decapoda. J Mar Biol Assoc United Kingdom, Brachyura, Varunidae). https://doi.org/10.1017/S002531541900002X
Landschoff J, Lackschewitz D, Kesy K, Reise K (2013) Globalization pressure and habitat change: Pacific rocky shore crabs invade armored shorelines in the Atlantic Wadden Sea. Aquat Invasions 8:77–87. https://doi.org/10.3391/ai.2013.8.1.09
Lawson Handley LJ, Estoup A, Evans DM, Thomas CE, Lombaert E, Facon B, Aebi A, Roy HE (2011) Ecological genetics of invasive alien species. Biocontrol 56:409–428. https://doi.org/10.1007/s10526-011-9386-2
Lee CE (2002) Evolutionary genetics of invasive species. Trends Ecol Evol 17:386–391. https://doi.org/10.1016/S0169-5347(02)02554-5
Lee S, Lee S-K, Rho HS, Kim W (2013) New Report of the Varunid Crabs, Hemigrapsus takanoi and Sestrostoma toriumii (Crustacea: Decapoda: Varunidae) from Korea. Anim Syst Evol Diversity 29:152–159. https://doi.org/10.5635/ASED.2013.29.2.152
Lefebvre A, Ellien C, Davoult D, Thiébaut E, Salomon JC (2003) Pelagic dispersal of the brittle-star Ophiothrix fragilis larvae in a megatidal area (English Channel, France) examined using an advection/diffusion model. Estuar Coast Shelf Sci 57:421–433. https://doi.org/10.1016/S0272-7714(02)00371-2
Lehnert SJ, DiBacco C, Jeffery NW, Blakeslee AMH, Isaksson J, Roman J, Wringe BF, Stanley RRE, Matheson K, McKenzie CH, Hamilton LC, Bradbury IR (2018) Temporal dynamics of genetic clines of invasive European green crab (Carcinus maenas) in eastern North America. Evol Appl 11:1656–1670. https://doi.org/10.1111/eva.12657
Lejeusne C, Saunier A, Petit N, Béguer M, Otani M, Carlton JT, Rico C, Green AJ (2014) High genetic diversity and absence of founder effects in a worldwide aquatic invader. Sci Rep 4:5808. https://doi.org/10.1038/srep05808
Leppäkoski E, Gollasch S, Gruszka P, Ojaveer H, Olenin S, Panov VE (2002) The Baltic – a sea of invaders. Can J Fish Aquat Sci 59:1175–1188. https://doi.org/10.1139/f02-089
Lockwood JL, Cassey P, Blackburn TM (2005) The role of propagule pressure in explaining species invasions. Trends Ecol Evol 20:223–228. https://doi.org/10.1016/j.tree.2005.02.004
Lord JP, Williams LM (2016) Increase in density of genetically diverse invasive Asian shore crab (Hemigrapsus sanguineus) populations in the Gulf of Maine. Biol Invasions 19:1153–1168. https://doi.org/10.1007/s10530-016-1304-1
Makino W, Miura O, Kaiser F, Geffray M, Katsube T, Urabe J (2018) Evidence of multiple introductions and genetic admixture of the Asian brush-clawed shore crab Hemigrapsus takanoi (Decapoda: Brachyura: Varunidae) along the Northern European coast. Biol Invasions 20:825–842. https://doi.org/10.1007/s10530-017-1604-0
Marin IN (2013) New data on the distribution of hairy-clawed shore crabs of the genus Hemigrapsus (Decapoda: Varunidae) along the Russian mainland coast of the Sea of Japan. Russ J Mar Biol 39:301–305. https://doi.org/10.1134/S106307401304007X
Markert A, Raupach MJ, Segelken-Voigt A, Wehrmann A (2014) Molecular identification and morphological characteristics of native and invasive Asian brush-clawed crabs (Crustacea: Brachyura) from Japanese and German coasts: Hemigrapsus penicillatus (De Haan, 1835) versus Hemigrapsus takanoi Asakura & Org Divers Evol. https://doi.org/10.1007/s13127-014-0176-4
Mingkid WM, Akiwa S, Watanabe S (2006) Morphological characteristics, pigmentation, and distribution of the sibling penicillate crabs, Hemigrapsus penicillatus (de Haan, 1835) and H. takanoi Asakura & Watanabe, 2005 (Decapoda, Brachyura, Grapsidae) in Tokyo Bay. Crustaceana 79:1107–1121
Moehler J, Wegner KM, Reise K, Jacobsen S (2011) Invasion genetics of Pacific oyster Crassostrea gigas shaped by aquaculture stocking practices. J Sea Res 66:256–262. https://doi.org/10.1016/j.seares.2011.08.004
Molnar JL, Gamboa RL, Revenga C, Spalding MD (2008) Assessing the global threat of invasive species to marine biodiversity. Front Ecol Environ 6:485–492. https://doi.org/10.1890/070064
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nijland R, Beekman J (2000) Hemigrapsus penicillatus de Haan 1835 waargenomen in Nederland. Het Zeepaard 60:169–171
Noël PY, Gruet Y (2008) Progression du crabe introduit Hemigrapsus takanoi Asukura & Watanabe 2005 (Crustacé, Décapode) vers le nord du Golfe de Gascogne. Bull la Société des Sci Nat l’Ouest la Fr Nouv Série 30:141–148
Noël PY, Tardy E, d’Udekem d’Acoz C (1997) Will the crab Hemigrapsus penicillatus invade the coasts of Europe? Comptes Rendus l’Académie des Sci Paris, Sci la Vie/Life Sci 320:741–745. https://doi.org/10.1016/S0764-4469(97)84823-8
O’Connor NJ (2014) Invasion dynamics on a temperate rocky shore: from early invasion to establishment of a marine invader. Biol Invasions 16:73–87. https://doi.org/10.1007/s10530-013-0504-1
Obert B, Herly M, Grotjahn M (2007) First records of two crabs from the North West Pacific Hemigrapsus sanguineus and H. takanoi at the coast of Lower Saxony. Germany Wadden Sea Newsl 1:21–22
Ojaveer H, Kotta J (2015) Ecosystem impacts of the widespread non-indigenous species in the Baltic Sea: literature survey evidences major limitations in knowledge. Hydrobiologia 750:171–185. https://doi.org/10.1007/s10750-014-2080-5
Paradis E (2010) Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Pérez-Portela R, Turon X, Bishop JDD (2012) Bottlenecks and loss of genetic diversity: Spatio-temporal patterns of genetic structure in an ascidian recently introduced in Europe. Mar Ecol Prog Ser 451:93–105. https://doi.org/10.3354/meps09560
Poux C, Gothland M, Holl AC, Spilmont N, Arnaud JF (2015) Highly polymorphic microsatellite markers in two species, the invasive shore crabs Hemigrapsus sanguineus and Hemigrapsus takanoi (Decapoda, Varunidae). Conserv Genet Resour 7:569–572. https://doi.org/10.1007/s12686-015-0426-6
Prevosti A, Ocaña J, Alonso G (1975) Distances between populations of Drosophila subobscura, based on chromosome arrangement frequencies. Theor Appl Genet 45:231–241. https://doi.org/10.1007/BF00831894
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2017) R: a language and environment for statistical computing.
Reusch TBH, Bolte S, Sparwel M, Moss AG, Javidpour J (2010) Microsatellites reveal origin and genetic diversity of Eurasian invasions by one of the world’s most notorious marine invader, Mnemiopsis leidyi (Ctenophora). Mol Ecol 19:2690–2699. https://doi.org/10.1111/j.1365-294X.2010.04701.x
Rius M, Darling JA (2014) How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol Evol 29:233–242. https://doi.org/10.1016/j.tree.2014.02.003
Rius M, Turon X, Bernardi G, Volckaert FAM, Viard F (2015) Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biol Invasions 17:869–885. https://doi.org/10.1007/s10530-014-0792-0
Rodgers PJ, Reaka ML, Hines AH (2011) A comparative analysis of sperm storage and mating strategies in five species of brachyuran crabs. Mar Biol 158:1733–1742. https://doi.org/10.1007/s00227-011-1687-6
Roman J (2006) Diluting the founder effect: cryptic invasions expand a marine invader’s range. Proc R Soc B 273:2453–2459. https://doi.org/10.1098/rspb.2006.3597
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:454–464. https://doi.org/10.1016/j.tree.2007.07.002
Rosenberg NA (2004) DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes 4:137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x
Rudnick D, Resh V (2005) Stable isotopes, mesocosms and gut content analysis demonstrate trophic differences in two invasive decapod crustacea. Freshw Biol 50:1323–1336. https://doi.org/10.1111/j.1365-2427.2005.01398.x
Ruiz GM, Fofonoff PW, Carlton JT, Wonham MJ, Hines AH (2000) Invasion of Coastal Marine Communities in North America: Apparent Patterns, Processes, and Biases. Annu Rev Ecol Syst 31:481–531. https://doi.org/10.2307/annurev.ecolsys.37.091305.30000016
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG (2001) The Population Biology of Invasive Species. Annu Rev Ecol Syst 32:305–332. https://doi.org/10.1146/annurev.ecolsys.32.081501.114037
Salomon JC, Breton M (1993) An atlas of long-term currents in the channel. Oceanol Acta 16:439–448
Seebens H, Gastner MT, Blasius B (2013) The risk of marine bioinvasion caused by global shipping. Ecol Lett 16:782–790. https://doi.org/10.1111/ele.12111
Simberloff D, Martin JL, Genovesi P, Maris V, Wardle DA, Aronson J, Courchamp F, Galil BS, García-Berthou E, Pascal M, Pyšek P, Sousa R, Tabacchi E, Vilà M (2013) Impacts of biological invasions: What’s what and the way forward. Trends Ecol Evol 28:58–66. https://doi.org/10.1016/j.tree.2012.07.013
Simon-Bouhet B, Garcia-Meunier P, Viard F (2006) Multiple introductions promote range expansion of the mollusc Cyclope neritea (Nassariidae) in France: evidence from mitochondrial sequence data. Mol Ecol 15:1699–1711. https://doi.org/10.1111/j.1365-294X.2006.02881.x
Svansson A (1975) Physical and chemical oceanography of the Skagerrak and the Kattegat.
Tepolt CK, Darling JA, Bagley MJ, Geller JB, Blum MJ, Grosholz ED (2009) European green crabs (Carcinus maenas) in the Northeastern Pacific: genetic evidence for high population connectivity and current-mediated expansion from a single introduced source population. Divers Distrib 15:997–1009. https://doi.org/10.1111/j.1472-4642.2009.00605.x
van den Brink AM, Wijnhoven S, McLay CL (2012) Competition and niche segregation following the arrival of Hemigrapsus takanoi in the formerly Carcinus maenas dominated Dutch delta. J Sea Res 73:126–136. https://doi.org/10.1016/j.seares.2012.07.006
van den Brink AM, Godschalk M, Smaal AC, Lindeboom H, McLay CL (2013) Some like it hot: the effect of temperature on brood development in the invasive crab Hemigrapsus takanoi (Decapoda: Brachyura: Varunidae). J Mar Biol Assoc UK 93:189–196. https://doi.org/10.1017/S0025315412000446
Viard F, Ellien C, Dupont L (2006) Dispersal ability and invasion success of Crepidula fornicata in a single gulf: insights from genetic markers and larval-dispersal model. Helgol Mar Res 60:144–152. https://doi.org/10.1007/s10152-006-0033-8
Wagner NK, Ochocki BM, Crawford KM, Compagnoni A, Miller TEX (2017) Genetic mixture of multiple source populations accelerates invasive range expansion. J Anim Ecol 86:21–34. https://doi.org/10.1111/1365-2656.12567
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184. https://doi.org/10.1007/s10592-005-9100-y
Waples RS, Do C (2008) LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756. https://doi.org/10.1111/j.1755-0998.2007.02061.x
Welles SR, Dlugosch KM (2018) Population Genomics of Colonization and Invasion. In: Rajora OP (ed) Population Genomics: Concepts, Approaches and Applications. Springer, Cham, pp 655–683
Winther NG, Johannessen JA (2006) North Sea circulation: Atlantic inflow and its destination. J Geophys Res Ocean 111:C12018. https://doi.org/10.1029/2005JC003310
Wolff WJ (2005) Non-indigenous marine and estuarine species in The Netherlands. Zool Meded Leiden 79:1–116
Wood CA, Bishop JDD, Davies CJ, Delduca EL, Hatton JC, Herbert RJH, Clark PF (2015) Hemigrapsus takanoi Asakura and Watanabe, 2005 (Crustacea: Decapoda: Brachyura: Grapsoidea): first records of the brush-clawed shore crab from Great Britain. BioInvasions Rec 4:109–113. https://doi.org/10.3391/bir.2015.4.2.07
Yamasaki I, Doi W, Mingkid WM, Yokota M, Strüssmann CA, Watanabe S (2011) Molecular-Based Method To Distinguish the Sibling Species Hemigrapsus penicillatus and Hemigrapsus takanoi (Decapoda: Brachyura: Varunidae). J Crustac Biol 31:577–581. https://doi.org/10.1651/10-3366.1
Acknowledgements
We thank A. Goedknegt, D. Thieltges, J. Sewell, P. Clark, J.P. Pezy and B. Lebreton for their efforts in collecting and sending crabs from The Netherlands, Great Britain and France, N. Weiler, S. Wagner and M. Bauer for their help with crab collections in Germany, and W. Wranik for sharing records and localities of the first H. takanoi in Wismar Bight. We thank J. Valqui for the introduction into the lab routines, and C. Schubart and his working group for invaluable help in optimising lab routines for crustacean population genetics and the very fruitful discussions during two stays of JCG at Regensburg University. Thanks also to T. Jombart for making the relaxing power of cat videos available during population genetics analyses via ‘showmekittens()’ in his great R package adegenet. We further thank the Institute of Clinical Molecular Biology in Kiel for providing Sanger sequencing as supported in part by the DFG Cluster of Excellence “Inflammation at Interfaces” and “Future Ocean”, and especially the technicians S. Greve, S. Arndt and T. Henke for technical support. We highly appreciate the comments of M. Rius and three anonymous reviewers which greatly helped to improve the manuscript. JCG is grateful for financial support by a PhD stipend from the German Environmental Foundation (DBU), project number 20014/335.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The study was in part funded by a PhD stipend of the German Environmental Foundation (DBU) awarded to JCG, project number 20014/335. All authors declare no conflict of interest.
Human and animal rights
This article does not contain any studies performed with human participants performed by any of the authors, nor were any experiments performed on captive animals. All applicable EU, national and/or institutional guidelines for the care and use of animals in research were followed. All procedures performed in the study involving animals were in accordance with the ethical standards of German law regarding maintaining animal welfare in research.
Additional information
Responsible Editor: M. Rius.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewed by N. Jeffery and undisclosed experts.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Geburzi, J.C., Ewers-Saucedo, C., Brandis, D. et al. Complex patterns of secondary spread without loss of genetic diversity in invasive populations of the Asian shore crab Hemigrapsus takanoi (Decapoda) along European coasts. Mar Biol 167, 180 (2020). https://doi.org/10.1007/s00227-020-03790-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-020-03790-y