Abstract
Key message
Two QTL with pleiotropic effects on plant height and spike length linked in coupling phase on chromosome 2DS were dissected, and diagnostic marker for each QTL was developed.
Abstract
Plant height (PHT) is a crucial trait related to plant architecture and yield potential, and dissection of its underlying genetic basis would help to improve the efficiency of designed breeding in wheat. Here, two quantitative trait loci (QTL) linked in coupling phase on the short arm of chromosome 2D with pleiotropic effects on PHT and spike length, QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, were separated and characterized. QPht/Sl.cau-2D.1 is a novel QTL located between SNP makers BS00022234_51 and BobWhite_rep_c63957_1472. QPht/Sl.cau-2D.2 is mapped between two SSR markers, SSR-2062 and Xgwm484, which are located on the same genomic interval as Rht8. Moreover, the diagnostic marker tightly linked with each QTL was developed for the haplotype analysis using diverse panels of wheat accessions. The frequency of the height-reduced allele of QPht/Sl.cau-2D.1 is much lower than that of QPht/Sl.cau-2D.2, suggesting that this novel QTL may be an attractive target for genetic improvement. Consistent with a previous study of Rht8, a significant difference in cell length was observed between the NILs of QPht/Sl.cau-2D.2. By contrast, there was no difference in cell length between NILs of QPht/Sl.cau-2D.1, indicating that the underlying molecular mechanism for these two QTL may be different. Collectively, these data provide a new example of QTL dissection, and the developed diagnostic markers will be useful in marker-assisted pyramiding of QPht/Sl.cau-2D.1 and/or QPht/Sl.cau-2D.2 with the other genes in wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat (Triticum aestivum L.) is an important food crop worldwide, providing calories and proteins consumed by humankind (Fischer et al. 2014; Shiferaw et al. 2013). Plant height (PHT) is a crucial trait related to plant architecture and yield potential (Cadalen et al. 1998; Peng et al. 1999; Sakamoto and Matsuoka 2004). Consequently, the use of dwarfing genes to reduce PHT and improve yield has been one of the main strategies in breeding modern high-yielding hexaploid bread wheat varieties. For example, during the green revolution, the introduction of semi-dwarf varieties into wheat (T. aestivum L.) appropriately reduced PHT and contributed significantly to a worldwide increase in potential grain yield (Peng et al. 1999). PHT is known to be typically under polygenic control (Bellucci et al. 2015; Tang et al. 2007). Thus, identification of QTL/gene controlling PHT would help to improve the efficiency of designed breeding in wheat.
To date, 24 genes influencing PHT have been identified and designated reduced height genes in wheat (McIntosh et al. 2017). Of these 24 major genes, Rht-B1b and Rht-D1b, which are located on chromosomes 4B and 4D, respectively (Pearce et al. 2011), were cloned and most widely used in wheat breeding (Borner et al. 2002; Cadalen et al. 1998). They encoded DELLA proteins, which are transcriptional regulators that reduce response to gibberellin. Thus, Rht-B1b and Rht-D1b belonged to the group of dwarfing genes that is insensitive to gibberellic acid (Pearce et al. 2011). The characterization of these two genes has enhanced our knowledge about PHT determination in wheat, and functional markers developed for them have been used in wheat breeding (Akman and Bruckner 2012; Borrell et al. 1991; Tang et al. 2009).
The other extensively used gene in wheat breeding is Rht8, which is derived from the Japanese variety Akakomugi and was introduced into Southern European wheat breeding in the 1930s by the Italian breeder Strampelli, together with the photoperiod-insensitive, early flowering Ppd-D1a allele (Lorenzetti 2000). Rht8 was mapped on the short arm of chromosome 2D, and a closely linked SSR marker named Xgwm261 was detected (Korzun et al. 1998). The 192-bp allele of Xgwm261 corresponds to a height-reducing phenotype of Rht8, attributing to a 7–8 cm reduction in PHT without pleiotropic effects on other agronomic traits (Korzun et al. 1998; Worland et al. 1998). Since then, the ‘diagnostic’ 192-bp allele was used to survey the presence of Rht8 in wheat cultivars (Ahmad and Sorrells 2002; Asplund et al. 2012; Bai et al. 2004; Chebotar et al. 2001; Liu et al. 2005; Worland et al. 2001a, b; Zhang et al. 2006). However, the 192-bp allele of Xgwm261 was not always linked to Rht8 (Ellis et al. 2007). A recent study showed that Rht8 is located in a genetic interval of 1.29 cM (DG279-DG371), which is 1.95 cM away from Xgwm261 (Gasperini et al. 2012). Thus, the development of a robust marker tightly linked to Rht8 is the priority for marker-assisted selection.
Over the past two decades, the successful application of quantitative–genetic methodology has facilitated identification of numerous QTL for PHT in wheat (Borner et al. 2002; Cadalen et al. 1998; Wurschum et al. 2015, 2017; Yu et al. 2014; Zanke et al. 2014). In our recent study, two QTL (QPht.cau-2D.1 and QPht.cau-2D.2) for PHT linked in coupling phase were mapped on the short arm of chromosome 2D (Zhai et al. 2016). Since both QTL are located on chromosome 2DS, this investigation was undertaken to determine the genetic relationship between QPht.cau-2D.1 and QPht.cau-2D.2 using segregating populations and NILs derived from a residual heterozygous line. Moreover, the diagnostic marker for each QTL was developed for marker-assisted selection in wheat breeding programmes. In addition, haplotype distribution of QPht.cau-2D.1 and QPht.cau-2D.2 in diverse panels of wheat accessions was also investigated.
Materials and methods
Plant materials
Following our previous study of QTL mapping using the recombinant inbred lines (RILs) of Yumai 8679 (Y8679)/Jing 411 (J411) (Zhai et al. 2016), SSR (simple sequence repeat) markers were further developed for linkage map construction and QTL analysis. In generation F9 of the RIL population, a RHL (RIL171) that carried the heterozygous segment at the genetic region from SSR markers SSR-2212 to SSR-2429 was self-pollinated to produce the F10 generation for further study (Fig. 1). The homozygotes (F10) without recombinant were selected as NILs (NILY8679 and NILJ411) to validate the presence of the QTL. Two plants that carried heterozygous segments covering the intervals from SSR-2212 to Xwmc503 and from Xcfd53 to SSR-2429 were identified to produce segregate populations (F11, population I and II) for genetic analysis (Fig. 2b). Then, the non-recombinant homozygotes of each population were selected and self-pollinated to produce NIL-I (F12, NIL-IY8679 and NIL-IJ411) and NIL-II (F12, NIL-IIY8679 and NIL-IIJ411).
Saturated genetic linkage map of chromosome 2D in the RIL population and the collinearity of the developed markers, Aegilops tauschii markers and corresponding physical position in the Chinese Spring RefSeq v1.0 sequence. The red segment means the heterozygous segment in RIL171 (color figure online)
Dissection of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2. a QTL mapping using a saturate genetic map. b Graphical genotypes of two populations (derived from RIL 171). c Performance of the members of two NIL pairs in three field trails. *** indicate significant differences at the 0.001 levels (Student’s t test)
In total, 1433 wheat accessions with varying ploidy were used to test the allele frequency of two diagnostic markers (STARP-2001 and SSR-2433). These included 1347 hexaploid wheat accessions (724 Chinese varieties/lines, 181 Chinese mini core accessions and 442 accessions from other countries (Supplementary Tables S1, S2 and S3, respectively), and 86 diploid accessions (Aegilops tauschii; Supplementary Table S4).
Field experiments
Populations I and II were tested in Hebei (37°56′N, 114°42′E). NILY8679 and NILJ411 were grown in two locations, Shanxi (36°08′N, 111°34′E) and Hebei, and NIL-I and NIL-II were grown in three locations, Beijing, Shanxi and Hebei (Supplementary Table S5). In each environment, two segregating populations were planted with a sowing rate of 20 seeds per row (1.5 m long and 0.3 m apart), while the NILs were planted 25 seeds per line. The irrigation and other management of all sites were in accordance with local standard practices.
Phenotypic evaluation and statistical analysis
Plant height (PHT), spike length (SL) and spikelet number (SN) of the individuals in populations I and II were measured before harvest. For the NILs, five plants from each line were used for phenotypic evaluation. PHT, SL, SN and length of 5 internodes from the main tillers were measured. For NILs, the mean values over three replications at each site were used for data analysis. Spikelet compactness (SC) was calculated by dividing the SN by the SL.
Statistical analysis was performed with IBM SPSS Statistics 20 (SPSS, Chicago, USA). Significance analysis was calculated using Student’s t test.
SSR marker development
Microsatellite markers were designed to construct the genetic map of chromosome 2DS. The reference sequence of wheat variety Chinese Spring (http://www.wheatgenome.org/) and Aegilops. tauschii genomic DNA contig sequence (http://aegilops.wheat.ucdavis.edu/ATGSP/) were extracted to design new SSR markers by using BLASTN method with the sequences of markers DG279 and DG371 (Gasperini et al. 2012) and the flanking sequence of the polymorphic SNPs in our previous results (Zhai et al. 2016). The PCR system included 5 μl 2 × Taq PCR StarMix, 2 μl primer (mixture of left and right primer), 2 μl DNA template (50–100 ng/μl) and 1 μl H2O. The PCR protocol was performed using PCR amplification as follows: 94 °C for 5 min; 36 cycles of 94 °C denaturation for 30 s, 55–57 °C primer annealing for 30 s, and 72 °C extending for 30 s; and finally, 72 °C for 10 min. For polymorphism detection, PCR products were separated using the method of 8% non-denaturing polyacrylamide gel electrophoresis (PAGE) (Marklund et al. 1995). The primers of 17 codominant SSR markers are listed in Supplementary Table S14.
Linkage map construction and QTL analysis
JoinMap 4.1 was used for constructing genetic linkage map of chromosome 2DS with LOD value above 5. A regression mapping algorithm (Stam 1993) and Kosambi’s mapping function (Kosambi 1943) were applied to determine the marker order and convert recombination into distances.
QTL mapping was determined with Windows QTL Cartographer 2.5 (Wang et al. 2006). Briefly, composite interval mapping (CIM) was performed using model 6 with forward and backward regression, five markers, and a 1-cM scanning window as cofactors. Empirical threshold LOD scores estimated with 1000 permutations at P ≤ 0.05 detected QTL with overlapping confidence intervals (± 2 LOD away from the peaks of likelihood ratios) were considered equivalent and named as suggested by McIntosh et al. (2017).
STARP marker development
To develop a diagnostic marker for QPht/Sl.cau-2D.1, the SNP marker AX-108988107 was converted into a semi-thermal asymmetric reverse marker (STARP) (Long et al. 2017). The primers included two asymmetrically modified AMAS primers (STARP-2001 F1 and STARP-2001 F2) and their same reverse primer (STARP-2001 R). STARP-2001 F1 was designed to amplify the Y8679 allele uniquely with 10 bp insertion (TGCTGACGAC) at 5′ terminus. Meanwhile, STARP-2001 F2 could amplify the J411 allele. The nucleotides substitution principle followed the suggestion of Long et al. (Long et al. 2017). The three primers were mixed in a ratio of 1:1:2 (STARP-2001 F1: STARP-2001 F2: STARP-2001 R) and diluted. A 10-μl PCR system containing 5 μl 2 × Taq PCR StarMix, 2 μl mixed primer, 2 μl DNA template (50–100 ng/μl) and 1 μl H2O was performed with 94 °C initial denaturation for 5 min, followed by 11 cycles of a 3-step touchdown PCR protocol starting at 94 °C for 30 s, then 66 °C for 30 s and 72 °C extending for 30 s, with the annealing temperature being decreased by 1 °C per cycle. This touchdown PCR protocol was accompanied with 30 cycles of 3 steps (94 °C for 30 s, then 55 °C for 30 s and 72 °C extending for 30 s) and then, finally, 72 °C for 10 min. Then, 8% PAGE was adopted to separate the length polymorphism.
Histological analysis
The medial sections of peduncle at flowering stage were collected from NIL-IY8679, NIL-IJ411, NIL-IIY8679 and NIL-IIJ411 plants, then fixed in FAA solution (50% (v/v) ethanol, 5% (v/v) glacial acetic acid and 4% (v/v) formaldehyde) and subjected to vacuum pumping for 30 min. Next, the internodes were dehydrated in a series of ethanol solutions (75% (v/v) ethanol, 85% (v/v) ethanol, 90% (v/v) ethanol, 95% (v/v) ethanol and anhydrous ethanol) and destained in a series of xylene solutions (3:1 ethanol: xylene, 1:1 ethanol: xylene, 1:3 ethanol: xylene, and pure xylene). The internodes were soaked in each ethanol and xylene solution for 1 h and then embedded in paraffin (He et al. 2017). Tissue sections were cut into 4-μm-thick slices, fixed on a glass slide, and stained with 1% sarranine and 0.5% fast green (G1031, http://www.servicebio.cn/).
Sequence analysis of candidate genes
Genomic DNA for Y8679 and J411 was used to build paired-end sequencing libraries with insert sizes of approximately 500 bp, according to vendor-provided instructions (Illumina). An average 5 × coverage of the assembled genome, with 150-bp paired-end reads for each accession, was generated with the Illumina HiSeq X Ten platform. All the sequence reads for each parent were mapped to the newly updated genome of Chinese Spring (RefSeq v1.0) in the Burrows–Wheeler Aligner program (BWA, ver. 0.7.15) with default parameters (Li and Durbin 2009). SNPs and InDels are identified by the HaplotypeCaller module. The sequence of each candidate gene including intron, exon, 3′-UTR region and 2-kb sequence upstream of translation start codon was used as query for analysis.
Results
Saturation of genetic Linkage map and QTL mapping of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2
To saturate the genetic linkage map for plant height (PHT) and spike length (SL) on the short arm of chromosome 2D, 632 SSR markers were developed, among which 17 markers exhibited polymorphism between parents Y8679 and J411 (Supplementary Table S14). The collinearity of these markers with Ae. tauschii markers was analysed, and the physical positions on the reference sequence of Chinese Spring are shown in Fig. 1. These 17 polymorphic SSR markers were used to genotype the 191 RIL lines for QTL analysis. The resulting genetic linkage map of chromosome 2D consisted of 75 markers spanning 70.11 cM in length (Supplementary Table S13). Consistent with our previous finding, two stable QTL in coupling phase controlling PHT (QPht.cau-2D.1 and QPht.cau-2D.2) were detected on chromosome 2DS by reanalysing the phenotypic data from 191 RIL lines in 9 individual environments and 1 combined analysis (BLUP) (Zhai et al. 2016). QPht.cau-2D.1 was located between SNP markers BS00022234_51 and BobWhite_rep_c63957_1472, which explained 4.39–11.94% of the phenotypic variation. QPht.cau-2D.2 was mapped to the genomic interval between SSR markers SSR-2062 and Xgwm484, which explained 4.12–12.96% of the phenotypic variations under different environments (Fig. 2a, Table 1). In the same positions of QTL for PHT, two major QTL for SL (QSl.cau-2D.1 and QSl.cau-2D.2) were also detected under all environments, which explained 30.94–40.63 and 31.31–41.95% of the phenotypic variations, respectively. All increasing alleles for PHT and SL came from J411 (Table 1). For simplification, hereafter, the two genomic regions covering PHT and SL were designated as QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, respectively.
Development of near isogenic lines harbouring QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2
To further evaluate the genetic effects of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2 on PHT and SL, one line (RIL171) from the F9 generation of the RIL population was selected for investigation because of its residual heterozygosity at the target QTL genomic regions. First, one pair of NILs (NILY8679 and NILJ411) from the self-pollinated progeny of RIL171 (F10) harbouring QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2 were genotyped using the wheat 660 K SNP array (https://wheat.pw.usda.gov/ggpages/topics/Wheat660_SNP_array_developed_by_CAAS.pdf). Of 630517 SNP markers with detected signal, only 434 (0.07%) exhibited polymorphism between NILY8679 and NILJ411, indicating that the genetic backgrounds of these two NILs had very high similarity (Supplementary Table S6). Notably, based on the information of SNP markers with genomic position, 154 of 434 polymorphic SNP markers (35.48%) were located on genomic interval flanking QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2 (Supplementary Table S7). Second, the progenies of the above NILs (F11) were used for phenotypic analysis. As shown in Table 2, significant differences were observed for SL, length of peduncle (PED) and length of 1st internode under peduncle (1IL) between NILY8679 and NILJ411 (P < 0.001) under different environments. By contrast, the differences in length of the second internode under peduncle (2IL), length of the third internode under peduncle (3IL) and length of the fourth internode under peduncle (4IL) were only detected under a specific environment. In addition, no difference was observed for spikelet number (SN). Consequently, PHT of NILJ411 was 12.62 and 15.08% (P < 0.001) higher than that of NILY8679 under two different environments. Correspondingly, spikelet compactness (SC) of NILY8679 was significantly (P < 0.001) higher than that of NILJ411 (Table 2).
Dissection of the two QTL linked in coupling phase
To further investigate the genetic effect of each QTL (QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2), eight polymorphic SSR markers between NILJ411 and NILY8679 were used to genotype the segregating populations of RIL 171 (F10). Two plants carrying different heterozygous genomic regions (I and II) were selected for further analysis, corresponding to QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, respectively (Fig. 2b). The single marker analysis for PHT, SL and SN was performed using the derived segregating populations I and II (F11). As expected, polymorphic SSR markers in each population were significantly (P < 0.001) associated with PHT and SL (Table 3). To further test the pleiotropic effects of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, two sets of NILs (F12, NIL-I and NIL-II) (Fig. 3) were developed from the two F11 populations. For PHT, SL and PED, NIL-IJ411 and NIL-IIJ411 (genotype same with J411) had significantly higher values than those of NIL-IY8679 and NIL-IIY8679 (genotype same with Y8679) under all environments, with average differences of 9.97% and 12.62, 16.25 and 19.56%, and 13.43% and 18.11%, respectively (Fig. 2c, Supplementary Table S9). In addition, the 1IL and 4IL of NIL-IIJ411 were also much higher (P < 0.001) than those of NIL-IIY8679 under all environments (Supplementary Table S9).
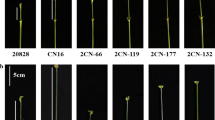
Culm and spike morphology of the NILs of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2 grown in Beijing (2016–2017 growing season). a Main tillers. Bars = 10 cm. b Spikes, peduncles and other internodes of NIL-IY8679 (left) and NIL-IJ411 (right). The bar represents 5 cm. c Spikes, peduncles and the internodes of NIL-IIY8679 (left) and NIL-IIJ411 (right). The bar represents 5 cm. PED, peduncle; 1I, first internode under peduncle; 2I, the second internode under peduncle; 3I, the third internode under peduncle; and 4I, the fourth internode under peduncle
Difference in PHT and SL between two alleles of STARP-2001 and SSR-2433 in the RIL population of five environments and one combined analysis (BLUP). The values represent the means (± SD) of RILs with the same genotype. *, **, *** indicate significant differences at the 0.05, 0.01, 0.001 levels (Student’s t test), respectively. STARP-2001-Y, the group with Y8679 type; STARP-2001-J, the group with J411 type; SSR-2433-Y, the group with Y8679 type; SSR-2433-J, the group with J411 type. The x-axis, five environments and one combined analysis(BLUP): E1, Beijing, 2010–2011; E3, Beijing, 2011–2012; E5, Anhui, 2012–2013; E7, Shaanxi, 2012–2013; E8, Beijing, 2014–2015; C indicates the combined QTL analysis based on the BLUP values across nine environments. a PHT between two alleles of STARP-2001. b SL between two alleles of STARP-2001. c PHT between two alleles of SSR-2433. d SL between two alleles of SSR-2433
Haplotype distributions of STARP-2001 and SSR-2433 in 724 common wheat varieties/lines from China (a, b) and 442 common wheat varieties/lines from other countries (c, d). I, Northern winter wheat region; II, Yellow and Huai River valley winter wheat region; III, low and middle Yangtze River valley winter wheat region; IV, south-western winter wheat region; V, southern winter wheat region; VI, north-eastern spring wheat region; VII, northern spring wheat region; VIII, north-western spring wheat region; IX, Qinghai–Tibet spring–winter wheat region; X, Xinjiang winter–spring wheat region
Effects of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2 on culm cell length
In wheat, internode elongation is caused by cell division and/or cell elongation (Chowdhry and Allan 1966; Gasperini et al. 2012). To investigate the underlying physiological bases of two QTL, longitudinal cell length in the median section of peduncle at the flowering stage in NILs of each QTL was measured. For QPht/Sl.cau-2D.1, NIL-IY8679 showed no difference in cells length compared to NIL-IJ411 (Fig. 6a, b and e). By contrast, there was significant difference in cell length between NIL-IIY8679 and NIL-IIJ411 of QPht/Sl.cau-2D.2. The cell length of NIL-IIJ411 with increasing allele was significantly longer than that of NIL-IIY8679 with decreasing allele for PHT (Fig. 6c, d, and f, P < 0.001).
Longitudinal culm sections of NILs of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2 from the flowering stage. Scanning micrographs of the medial zone of the fully elongated peduncle in a NIL-IY8679, b NIL-IJ411, c NIL-IIY8679 and d NIL-IIJ411. Bars = 100 μm. Comparisons of parenchymatic cell length (μm) in medial sections of the peduncle from e NILs of QPht/Sl.cau-2D.1 and fQPht/Sl.cau-2D.2. NS, no significance P = 0.05; ***, t test P < 0.001. Bars represent the standard deviation
Haplotype analysis of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2
To analyse the haplotype distribution of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, one SNP maker (AX-108988107) was converted into convenient STARP marker (STARP-2001) (Fig. 7a), which was used as the diagnostic marker of QPht/Sl.cau-2D.1. In addition, SSR-2433 (Fig. 7b) was selected as the diagnostic marker for QPht/Sl.cau-2D.2. To test the efficiency of these two markers, the 191 RIL populations were genotyped and analysed. As expected, the group of STARP-2001-J allele had higher PHT and longer SL than that of STARP-2001-Y allele, with differences of 3.3–5.86 and 10.65–13.87% (P < 0.05) under different environments (Fig. 4a, b). Similarly, PHT and SL of the SSR-2433-J allele were much higher (P < 0.001) than those of the SSR-2433-Y allele (5.10–7.07 and 12.37–15.73%) (Fig. 4c, d). Collectively, our data exhibited that the alleles of STARP-2001-Y and SSR-2433-Y were significantly associated with the height-reduced alleles of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, respectively.
To further explore implications of our findings on wheat breeding, the haplotype of 724 common wheat varieties/lines from China was first analysed using diagnostic markers STARP-2001 and SSR-2433. As shown in Fig. 5, both the STARP-2001-Y and SSR-2433-Y alleles were present in 6 of 10 agro-ecological production zones, including I, II, III, IV, VIII and X. The frequency of STARP-2001-Y was highest in zone II (45.60%), followed by zones III (33.33%), VIII (33.33%), IV (31.71%), I (23.26%) and X (1.83%) (Fig. 5a, Supplementary Table S11). The frequency of SSR-2433-Y allele in different zones was in the order of II (86.67%) > X (78.05%) > IV (51.22%) > III (48.15%) > I (46.51%) > VIII (33.33%) (Fig. 5b, Supplementary Table S11). When comparing the alleles of STARP-2001-Y and SSR-2433-Y in the same zone, the frequency of SSR-2433-Y was much higher than that of STARP-2001-Y in zones I, II, III, IV and X, especially in the major wheat production zone X (Fig. 5a, b, Supplementary Table S11). Furthermore, 442 common wheat varieties/lines from other countries were tested by the two diagnostic markers. The STARP-2001-Y allele was present in 48 of 442 accessions (10.86%), spreading in five continents, including Asia, Europe, North America, South America and Oceania. The SSR-2433-Y allele was detected in 63 of 442 accessions (14.25%), which are derived from Asia, Europe, Africa and North America (Fig. 5c, d, Supplementary Table S12).
To trace the origin of dwarf genes in the QTL intervals of QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2, mini core common wheat collections in China with 181 accessions were further genotyped using diagnostic markers STARP-2001 and SSR-2433. The STARP-2001-Y allele was detected in 20 of 181 accessions (11.04%). By contrast, SSR-2433-Y allele presented in 73 of 181 accessions (40.33%) (Supplementary Table S2). In addition, a diverse panel of 86 accessions of Aegilops tauschii, the D-genome progenitor of T. aestivum, was also analysed. The results exhibited that the STARP-2001-Y allele was not detected in 86 accessions of Aegilops tauschii, but the SSR-2433-Y allele presented in 39 of 86 accessions (45.35%) (Supplementary Table S4).
Sequence variations in candidate genes for QPht/Sl.cau-2D.1 and Rht8
To identify the candidate genes for QPht/Sl.cau-2D.1 and Rht8, we analysed the gene models in the mapping interval of the Chinese Spring RefSeq v.1.0 sequence. In rice, WRKY transcription factor Dlf1 and gibberellin-inactivating 2-beta-dioxygenase gene played important roles in the regulation of PHT (Cai et al. 2014; Lo et al. 2008). Interestingly, of 38 predicted genes in the 2.5-Mb interval of QPht/Sl.cau-2D.1 (BS00022234_51-BobWhite_rep_c63957_1472), TraesCS2D01G051000 and TraesCS2D01G051500 encoded two WRKY transcription factors, and TraesCS2D01G049700 encoded gibberellin 2-beta-dioxygenase, which may be the candidate genes (Supplementary Table S10). DNA sequence analysis showed that there were 5 SNPs and 3 InDels between TraesCS2D01G051500 of parental lines Y8679 and J411, including 3 SNPs in exon 1, 1 InDel in 3′-UTR region, 1 InDel in intron 2 and 2 SNPs and 1 InDel in the upstream region of translation start codon (Fig. 8, Supplementary Table S15). However, no sequence difference between Y8679 and J411 was detected for TraesCS2D01G049700 and TraesCS2D01G051000.
Structure of candidate genes showing the nucleotide and amino acid sequences polymorphism between Y8679 and J411. Lines, blue boxes and white boxes represent introns, exons and untranslated regions in the gene, respectively. Nucleotide and amino acid sequences of Y8679 and J411 are shown in red and black font, respectively. The numbers in bracket represent the positions of nucleotide or amino acid sequences relative to ATG. – represents deletion. F.S. indicates frame shift. a Structure of TraesCS2D01G051500. b Structure of TraesCS2D01G055700. c Structure of TraesCS2D01G058700 (color figure online)
The well-known reduced height genes Rht-B1 and Rht-D1 in wheat and several plant height related genes in rice (DLT, SMOS1/SHB and SLR1) encoded transcriptional factors belong to the GRAS family (De Vleesschauwer et al. 2016; Qiao et al. 2017; Tong et al. 2009). In rice, OsCCC1 was involved in cell elongation by regulating ion (Cl−, K+, and Na+) homoeostasis to maintain cellular osmotic potential, which affected PHT in turn (Chen et al. 2016). Based on the new reference sequence of Chinese Spring (RefSeq v.1.0), the mapped region of Rht8 between SSR-2062 and Xgwm484 contained 419 predicted genes, among which TraesCS2D01G055700 and TraesCS2D01G058700 encoded two GRAS transcription factors, and TraesCS2D01G059300 encoded a cation–chloride cotransporter, which may be the candidate genes (Supplementary Table S10). DNA sequence analysis revealed that there were 38 SNPs and 8 InDels between TraesCS2D01G055700 of Y8679 and J411, 3 SNPs and 3 InDels in the exon, and 35 SNPs and 5 InDels in the upstream region of translation start codon. For TraesCS2D01G058700, only 2 SNP variants were found, which were located in intron 4 and the upstream region of translation start codon, respectively (Fig. 8, Supplementary Table S15). By contrast, the sequence of TraesCS2D01G095300 in Y8679 was identical to that in J411.
Discussions
Two QTL for PHT/SL linked in coupling phase on chromosomes 2DS
Neighbouring QTL that are linked in coupling phase are commonly observed in primary QTL analysis (Fan et al. 2017; Wu et al. 2015). To date, many studies have tried to dissect QTL in coupling phase using NILs or residual heterozygous lines and found that coupling QTL were partially attributed to tightly linked independent QTL (Chemayek et al. 2017; Shen et al. 2011). For example, Wu et al. dissected two tightly linked QTL for PHT (qPH3) and yield per plant (qYD3) by using two rice near isogenic populations (Wu et al. 2015). In wheat, tight repulsion linkage between Sr36 and Sr39 was revealed by genetic, cytogenetic and molecular analyses (Chemayek et al. 2017). Recently, using 191 RILs derived from Y8679 and J411, two QTL (QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2) controlling PHT and SL were mapped on the short arm of chromosome 2D. However, it is difficult to conclude whether QPht/Sl.cau-2D.1 is a shadow or genuine QTL because it is only 7 cM away from QPht/Sl.cau-2D.2 (Zhai et al. 2016). Here, we dissected these two QTL (QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2) using segregating populations and NILs derived from a residual heterozygous line (RIL171). They were linked in coupling phase on chromosome 2DS, with increased alleles from the same parent (J411). QPht/Sl.cau-2D.1 was located between SNP markers BS00022234_51 and BobWhite_rep_c63957_1472, and QPht/Sl.cau-2D.2 was located between SSR markers SSR-2062 and Xgwm484. The results of NILs indicated that the genetic effect of these two QTL is similar, which could reduce plant height by 4.79–14.32 and 9.49–14.43 cm under different environments (Table 2). Our present study provides a new example of QTL dissection in wheat, but the underlying molecular basis of two QTL (QPht/Sl.cau-2D.1 and QPht/Sl.cau-2D.2) is still an area for further elucidation.
Robust diagnostic SSR marker for QPht/Sl.cau-2D.2
Numerous studies exhibited that there were major QTL/genes on chromosome 2DS, such as the well-known and widely used dwarf gene Rht8 (Chebotar et al. 2013; Griffiths et al. 2012; Zanke et al. 2014). Recently, using a fine-resolution mapping approach, the Rht8 genetic interval has been reduced from 20.5 to 1.29 cM, flanking by DG279 and DG371 (Gasperini et al. 2012). Comparative analysis revealed that QPht/Sl.cau-2D.2 of the present study is located on the same genetic interval of Rht8 (Gasperini et al. 2012). Moreover, consistent with the effect of Rht8 on culm cell elongation, significant difference in cell length was also observed between NILs of QPht/Sl.cau-2D.2 (Fig. 6c, d). This, we speculated that QPht/Sl.cau-2D.2 may be the well-characterized Rht8 gene.
Due to the widespread use of Rht8 in wheat breeding, assessing the distribution of height-reducing alleles at the Rht8 is of interest for breeding purposes. Following the identification of Xgwm261, a closely linked marker to Rht8 with the 192-bp allele corresponding to a height-reducing phenotype, the gwm261192bp allele has been taken as diagnostic marker for Rht8 (Ahmad and Sorrells 2002; Bai et al. 2004; Chebotar et al. 2001; Liu et al. 2005; Zhang et al. 2006). However, several instances were found in which the 192-bp allele was not associated with a height-reducing phenotype (Ellis et al. 2007). For example, a source variety of the Green Revolution semi-dwarfing genes, Norin10, carrying a 192-bp allele of Xgwm261, was independent of the height reduction effect of Rht8 (Ellis et al. 2007). Consistent with this, our data revealed that Xgwm261 was located in the genetic interval of QPht/Sl.cau-2D.1, instead of QPht/Sl.cau-2D.2. Thus, we developed a codominant SSR marker (SSR-2433) that was tightly linked to QPht/Sl.cau-2D.2/Rht8. Moreover, the SSR-2433 locus only has two allelic variants in the diversity panel of wheat accessions, which may be an ideal diagnostic marker for the QPht/Sl.cau-2D.2/Rht8 gene.
A novel QTL for PHT/SL tightly linked to dwarf gene Rht8
Previous studies exhibited that numerous QTL/genes controlling PHT were detected in wheat (Borner et al. 2002; Gao et al. 2015a, b; Peng et al. 2003; Quarrie et al. 2005; Singh et al. 2016; Tian et al. 2017). Specifically, 24 dwarfing genes (Rht1–Rht24) have been catalogued in wheat (McIntosh et al. 2017). However, quite a few genes for reduced stature have been used in wheat breeding, because most showed strongly negative effects on grain yield (Chapman et al. 2007; Zhang et al. 2013a). Thus, it is necessary to explore and utilize new QTL/genes controlling PHT. For example, the new reduced PHT gene Rht24 was important and extensively used in wheat breeding programmes (Wurschum et al. 2017). In the present and our previous studies, we identified a novel QTL for PHT neighbouring to Rht8, QPht/Sl.cau-2D.1, by the following evidences: (1) QPht/Sl.cau-2D.1 was mapped to a position distal to Rht8 on the short arm of chromosome 2D (Fig. 2a, Table 1); (2) QPht/Sl.cau-2D.1 had no significant effect on culm cell elongation, which is obviously different from that of Rht8 (Fig. 6).
Common wheat has an allohexaploid genome structure, which was hybrid by T. turgidum (AABB) and Aegilops tauschii (DD) 0.43 million years ago (International Wheat Genome Sequencing 2014; Petersen et al. 2006) To investigate the origin of the novel QPht/Sl.cau-2D.1 during wheat evolution, we analysed the allelic variation in the diagnostic marker STARP-2001 in a diverse panel of 86 accessions of Aegilops tauschii., but the STARP-2001-Y allele was not detected, which is significantly associated with the height-reduced allele of QPht/Sl.cau-2D.1. Thus, it seems that the novel height-reduced allele of QPht/Sl.cau-2D.1 was generated after the formation of allohexaploid wheat. Notably, haplotype analysis revealed that the height-reduced allele of QPht/Sl.cau-2D.1 has not been widely used in wheat breeding compared to Rht8. Thus, QPht/Sl.cau-2D.1 may be a favourable QTL in the genetic improvement in PHT in wheat, but the effect of different alleles of QPht/Sl.cau-2D.1 on grain yield and their potential utilization needs further investigation.
Candidate genes of Rht8 and QPht/Sl.cau-2D.1
To date, several genes controlled PHT have been cloned in wheat, such as Rht-B1, Rht-D1, TaSTE and GA2oxA9 (Rht18) (Ford et al. 2018; Gasperini et al. 2012; Peng et al. 1999; Zhang et al. 2013b). Specifically, Rht-B1 and Rht-D1 encoded transcriptional factors belong to the GRAS family (Peng et al. 1999; Sun 2010). Different alleles of Rht-B1 and Rht-D1 have been also identified, which produce dwarfs with a broad range of plant height (Pearce et al. 2011). Semi-dwarfing alleles Rht-B1d and Rht-B1e introduce premature stop codons within the amino-terminal coding region, whereas the severe dwarfism of Rht-B1c gene is caused by an intragenic insertion of 90 bp within the highly conserved amino-terminal DELLA domain (Pearce et al. 2011). In the present study, we found that two of the three candidate genes for Rht8 (TraesCS2D01G055700 and TraesCS2D01G058700) encoded GRAS transcription factors. Notably, a 35-bp deletion in the coding region of TraesCS2D01G055700 was found in parental line J411 as compared to Y8679, which resulted in frame shift for translation. Moreover, another two SNP variations between Y8679 and J411 leaded to amino acid substitutions. In addition, 35 SNPs and 5 InDels in the upstream region of translation start codon were detected (Fig. 8, Supplementary Table S15). Collectively, we proposed that the role of TraesCS2D01G055700 in plant height merit for further investigation.
In rice, Dlf1, a WRKY transcription factor, acts as a transactivator to downregulate Ehd2/RID1/OsId1 in the signal transduction pathway of flowering and plays an important role in the regulation of PHT (Cai et al. 2014). Of the three candidate genes for QPht/Sl.cau-2D.1, TraesCS2D01G051000 and TraesCS2D01G051500 encoded two WRKY transcription factors, but only TraesCS2D01G051500 has sequence variations between Y8679 and J411, including one SNP (A/C) in exon 1 that leaded to the substitution of 1 amino acid (I/L) and 2 SNPs and 1 InDel in the upstream region of translation start codon (Fig. 8, Supplementary Table S15). Therefore, detailed studies would be necessary to analyse the effect of these variations in the function of TraesCS2D01G051500 and plant height.
Epigenetics regulate gene expression without effects on gene sequences, which was involved in diverse aspects of plant development (Bender 2002; Heer et al. 2018; Paszkowski and Mittelsten Scheid 1998). The repression of FLC expression in Arabidopsis by vernalization is one of the well-studied examples of the epigenetic regulation of a gene in plants (Bastow et al. 2004; Questa et al. 2016). Recently, Zhang et al.(2012) identify a DNA hypomethylation epi-allele in rice and demonstrate that repression of FIE1 via DNA methylation and H3K9me2 is essential for plant height. Thereby, although the sequences of TraesCS2D01G049700, TraesCS2D01G051000 and TraesCS2D01G095300 in Y8679 were identical to those in J411, these genes may also be the candidate genes and further experiment is needed to validate which one is the real gene for Rht8 and QPht/Sl.cau-2D.1.
In conclusion, two QTL with pleiotropic effects on plant height and spike length linked in coupling phase on chromosome 2DS in common wheat were separated, and the diagnostic marker tightly linked with each QTL was developed, which corresponded to a novel QTL QPht/Sl.cau-2D.1 and well-known Rht8 gene, respectively. Of six candidate genes for two QTL, three have sequence variations between parental lines Y8679 and J411. Remarkably, the frequency of the height-reduced allele of QPht/Sl.cau-2D.1 in diverse panels of wheat accessions is much lower than that of QPht/Sl.cau-2D.2. Taken together, these data provide a new example of QTL dissection, and QPht/Sl.cau-2D.1 may be an attractive target for genetic improvement in wheat breeding.
Author contribution statement
ZN and AZ conceived the project; LC, ZC and RB carried out experiments; XC and HZ participated in field trials; LC, ZC and XC performed marker development of the QTL region of interest; LC and ZH developed the segregation populations and near isogenic lines; LC, ZC and RB performed phenotyping of the segregation populations and the near isogenic lines; QS, HP, YY, ZH, MX and WG assisted in revising the manuscript; LC analysed experimental results and wrote the manuscript.
Change history
24 September 2019
This article was unintentionally published twice in this journal.
24 September 2019
Theoretical and Applied Genetics
References
Ahmad M, Sorrells ME (2002) Distribution of microsatellite alleles linked to Rht8 dwarfing gene in wheat. Euphytica 123:235e–240e. https://doi.org/10.1023/A:1014962016967
Akman H, Bruckner P (2012) Marker assisted selection for Rht8 and Rht-D1b dwarfing genes in winter wheat breeding program. New Biotechnol 29:S139e. https://doi.org/10.1016/j.nbt.2012.08.389
Asplund L, Leino MW, Hagenblad J (2012) Allelic variation at the Rht8 locus in a 19th century wheat collection. Sci World J. https://doi.org/10.1100/2012/385610
Bai GH, Das MK, Carver BF, Xu XY, Krenzer EG (2004) Covariation for microsatellite marker alleles associated with Rht8 and coleoptile length in winter wheat. Crop Sci 44:1187e–1194e. https://doi.org/10.2135/cropsci2004.1187
Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427:164e–167e. https://doi.org/10.1038/nature02269
Bellucci A, Torp AM, Bruun S, Magid J, Andersen SB, Rasmussen SK (2015) Association mapping in scandinavian winter wheat for yield, plant height, and traits important for second-generation bioethanol production. Front Plant Sci. https://doi.org/10.3389/fpls.2015.01046
Bender J (2002) Plant epigenetics. Curr Biol CB 12:R412e–R414e
Borner A, Schumann E, Furste A, Coster H, Leithold B, Roder S, Weber E (2002) Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor Appl Genet 105:921e–936e. https://doi.org/10.1007/s00122-002-0994-1
Borrell AK, Incoll LD, Dalling MJ (1991) The influence of the Rht1 and Rht2 alleles on the growth of wheat stems and ears. Ann Bot Lond 67:103e–110e. https://doi.org/10.1093/oxfordjournals.aob.a088108
Cadalen T, Sourdille P, Charmet G, Tixier MH, Gay G, Boeuf C, Bernard S, Leroy P, Bernard M (1998) Molecular markers linked to genes affecting plant height in wheat using a doubled-haploid population. Theor Appl Genet 96:933e–940e. https://doi.org/10.1007/s001220050823
Cai Y, Chen X, Xie K, Xing Q, Wu Y, Li J, Du C, Sun Z, Guo Z (2014) Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 9:e102529e. https://doi.org/10.1371/journal.pone.0102529
Chapman SC, Mathews KL, Trethowan RM, Singh RP (2007) Relationships between height and yield in near-isogenic spring wheats that contrast for major reduced height genes. Euphytica 157:391e–397e. https://doi.org/10.1007/s10681-006-9304-3
Chebotar SV, Korzun VN, Sivolap YM (2001) Allele distribution at locus WMS261 marking the dwarfing gene Rht8 in common wheat cultivars of southern Ukraine. Russ J Genet+ 37:894e–898e. https://doi.org/10.1023/a:1016773515628
Chebotar GO, Chebotar SV, Motsnyy II, Sivolap YM (2013) Clarification of the Rht8-Ppd-D1 gene linkage on the 2D chromosome of winter bread wheat. Cytol Genet+ 47:70e–74e. https://doi.org/10.3103/s0095452713020047
Chemayek B, Bansal UK, Qureshi N, Zhang P, Wagoire WW, Bariana HS (2017) Tight repulsion linkage between Sr36 and Sr39 was revealed by genetic, cytogenetic and molecular analyses. Theor Appl Genet 130:587e–595e. https://doi.org/10.1007/s00122-016-2837-5
Chen ZC, Yamaji N, Fujii-Kashino M, Ma JF (2016) A cation-chloride cotransporter gene is required for cell elongation and osmoregulation in rice. Plant Physiol 171:494e–507e. https://doi.org/10.1104/pp.16.00017
Chowdhry AR, Allan RE (1966) Culm length and differential development of the coleoptile, root, and subcrown internode of near-isogenic wheat lines1. Crop Sci 6:49e–51e. https://doi.org/10.2135/cropsci1966.0011183X000600010015x
De Vleesschauwer D, Seifi HS, Filipe O, Haeck A, Huu SN, Demeestere K, Hofte M (2016) The DELLA protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice. Plant Physiol 170:1831e–1847e. https://doi.org/10.1104/pp.15.01515
Ellis MH, Bonnett DG, Rebetzke GJ (2007) A 192 bp allele at the Xgwm261 locus is not always associated with the Rht8 dwarfing gene in wheat (Triticum aestivum L.). Euphytica 157:209e–214e. https://doi.org/10.1007/s10681-007-9413-7
Fan C, Zhai H, Wang H, Yue Y, Zhang M, Li J, Wen S, Guo G, Zeng Y, Ni Z, You M (2017) Identification of QTLs controlling grain protein concentration using a high-density SNP and SSR linkage map in barley (Hordeum vulgare L.). BMC Plant Biol 17:122e. https://doi.org/10.1186/s12870-017-1067-6
Fischer T, Byerlee D, Edmeades G (2014) Crop yields and global food security: will yield increase continue to feed the world? Aciar Monogr Ser 158:1e–634e. https://doi.org/10.1017/S0021859614000884
Ford B, Foo E, Sharwood RE, Karafiatova M, Vrána J, MacMillan C, Nichols DS, Steuernagel B, Uauy C, Doležel J, Chandler P, Spielmeyer W (2018) Rht18 semi-dwarfism in wheat is due to increased expression of GA 2-oxidaseA9 and lower GA content. Plant Physiol. https://doi.org/10.1104/pp.18.00023
Gao F, Wen W, Liu J, Rasheed A, Yin G, Xia X, Wu X, He Z (2015a) Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat Cross Zhou 8425B/Chinese Spring. Front Plant Sci 6:1099e. https://doi.org/10.3389/fpls.2015.01099
Gao FM, Wen WE, Liu JD, Rasheed A, Yin GH, Xia XC, Wu XX, He ZH (2015b) Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring. Front Plant Sci 9:1101. https://doi.org/10.3389/fpls.2015.01099
Gasperini D, Greenland A, Hedden P, Dreos R, Harwood W, Griffiths S (2012) Genetic and physiological analysis of Rht8 in bread wheat: an alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J Exp Bot 63:6057. https://doi.org/10.1093/jxb/ers292
Griffiths S, Simmonds J, Leverington M, Wang Y, Fish L, Sayers L, Alibert L, Orford S, Wingen L, Snape J (2012) Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol Breed 29:159e–171e. https://doi.org/10.1007/s11032-010-9534-x
He Z, Zeng J, Ren Y, Chen D, Li W, Gao F, Cao Y, Luo T, Yuan G, Wu X, Liang Y, Deng Q, Wang S, Zheng A, Zhu J, Liu H, Wang L, Li P, Li S (2017) OsGIF1 positively regulates the sizes of stems, leaves, and grains in rice. Front Plant Sci 8e:1730e. https://doi.org/10.3389/fpls.2017.01730
Heer K, Mounger J, Boquete MT, Richards CL, Opgenoorth L (2018) The diversifying field of plant epigenetics. New Phytol 217:988e–992e. https://doi.org/10.1111/nph.14985
International Wheat Genome Sequencing C (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788e. https://doi.org/10.1126/science.1251788
Korzun V, Roder MS, Ganal MW, Worland AJ, Law CN (1998) Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor Appl Genet 96:1104e–1109e. https://doi.org/10.1007/s001220050845
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugenic 12:172e–175e. https://doi.org/10.1111/j.1469-1809.1943.tb02321.x
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754e–1760e. https://doi.org/10.1093/bioinformatics/btp324
Liu Y, Liu DC, Zhang HY, Wang J, Sun JZ, Guo XL, Zhang AM (2005) Allelic variation, sequence determination and microsatellite screening at the XGWM261 locus in Chinese hexaploid wheat (Triticum aestivum) varieties. Euphytica 145:103e–112e. https://doi.org/10.1007/s10681-005-0549-z
Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20:2603e–2618e. https://doi.org/10.1105/tpc.108.060913
Long YM, Chao WS, Ma GJ, Xu SS, Qi LL (2017) An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor Appl Genet 130:597e–607e. https://doi.org/10.1007/s00122-016-2838-4
Lorenzetti R (2000) The wheat science. The green revolution of Nazareno Strampelli. J Genet Breed (special publication)
Marklund S, Chaudhary R, Marklund L, Sandberg K, Andersson L (1995) Extensive mtDNA diversity in horses revealed by PCR-SSCP analysis. Anim Genet 26:193e–196e
McIntosh R, Rogers J, Dubcovsky J, Morris C, Xia XC (2017) Catalogue of gene symbols for wheat: 2017 supplement
Paszkowski J, Mittelsten Scheid O (1998) Plant genes: the genetics of epigenetics. Curr Biol CB 8:R206e–R208e
Pearce S, Saville R, Vaughan SP, Chandler PM, Wilhelm EP, Sparks CA, Al-Kaff N, Korolev A, Boulton MI, Phillips AL, Hedden P, Nicholson P, Thomas SG (2011) Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol 157:1820e–1831e. https://doi.org/10.1104/pp.111.183657
Peng JR, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400:256e–261e. https://doi.org/10.1038/22307
Peng J, Ronin Y, Fahima T, Roder MS, Li Y, Nevo E, Korol A (2003) Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc Natl Acad Sci U S A 100:2489e–2494e. https://doi.org/10.1073/pnas.252763199
Petersen G, Seberg O, Yde M, Berthelsen K (2006) Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol Phylogenet Evol 39:70e–82e. https://doi.org/10.1016/j.ympev.2006.01.023
Qiao S, Sun S, Wang L, Wu Z, Li C, Li X, Wang T, Leng L, Tian W, Lu T, Wang X (2017) The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell 29:292e–309e. https://doi.org/10.1105/tpc.16.00611
Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusic D, Waterman E, Weyen J, Schondelmaier J, Habash DZ, Farmer P, Saker L, Clarkson DT, Abugalieva A, Yessimbekova M, Turuspekov Y, Abugalieva S, Tuberosa R, Sanguineti MC, Hollington PA, Aragues R, Royo A, Dodig D (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring x SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110:865e–880e. https://doi.org/10.1007/s00122-004-1902-7
Questa JI, Song J, Geraldo N, An H, Dean C (2016) Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353:485e–488e. https://doi.org/10.1126/science.aaf7354
Sakamoto T, Matsuoka M (2004) Generating high-yielding varieties by genetic manipulation of plant architecture. Curr Opin Biotechnol 15:144e–147e. https://doi.org/10.1016/j.copbio.2004.02.003
Shen B, Yu WD, Du JH, Fan YY, Wu JR, Zhuang JY (2011) Validation and dissection of quantitative trait loci for leaf traits in interval RM4923-RM402 on the short arm of rice chromosome 6. J Genet 90:39e–44e. https://doi.org/10.1007/s12041-011-0019-4
Shiferaw B, Smale M, Braun HJ, Duveiller E, Reynolds M, Muricho G (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur 5:291e–317e. https://doi.org/10.1007/s12571-013-0263-y
Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Kumar S, Campbell HL (2016) Genetic mapping of common bunt resistance and plant height QTL in wheat. Theor Appl Genet 129:243e–256e. https://doi.org/10.1007/s00122-015-2624-8
Stam P (1993) Construction of integrated genetic-linkage maps by means of a new computer package: joinmap. Plant Journal 3:739e–744e. https://doi.org/10.1111/j.1365-313X.1993.00739.x
Sun TP (2010) Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol 154:567e–570e. https://doi.org/10.1104/pp.110.161554
Tang J, Teng W, Yan J, Ma X, Meng Y, Dai J, Li J (2007) Genetic dissection of plant height by molecular markers using a population of recombinant inbred lines in maize. Euphytica 155:117e–124e. https://doi.org/10.1007/s10681-006-9312-3
Tang N, Jiang Y, He BR, Hu YG (2009) The effects of dwarfing genes (Rht-B1b, Rht-D1b, and Rht8) with different sensitivity to GA(3) on the coleoptile length and plant height of wheat. Agr Sci China 8:1028e–1038e. https://doi.org/10.1016/S1671-2927(08)60310-7
Tian X, Wen W, Xie L, Fu L, Xu D, Fu C, Wang D, Chen X, Xia X, Chen Q, He Z, Cao S (2017) Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front Plant Sci 8:1379e. https://doi.org/10.3389/fpls.2017.01379
Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J 58:803e–816e. https://doi.org/10.1111/j.1365-313X.2009.03825.x
Wang S, Basten CJ, Zeng Z-B (2006) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh
Worland AJ, Korzun V, Roder MS, Ganal MW, Law CN (1998) Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theor Appl Genet 96:1110e–1120e. https://doi.org/10.1007/s001220050846
Worland AJ, Sayers EJ, Korzun V (2001a) Allelic variation at the dwarfing gene Rht8 locus and its significance in international breeding programmes. Euphytica 119:157e–161e. https://doi.org/10.1023/a:1017582122775
Worland AJ, Sayers EJ, Korzun V (2001b) Allelic variation at the dwarfing gene RHT8 locus and its significance in international breeding programmes. Dev Plant Breed 9:747e–753e
Wu B, Mao DH, Liu TM, Li ZX, Xing YZ (2015) Two quantitative trait loci for grain yield and plant height on chromosome 3 are tightly linked in coupling phase in rice. Mol Breed. https://doi.org/10.1007/s11032-015-0345-y
Wurschum T, Langer SM, Longin CFH (2015) Genetic control of plant height in European winter wheat cultivars. Theor Appl Genet 128:865e–874e. https://doi.org/10.1007/s00122-015-2476-2
Wurschum T, Langer SM, Longin CFH, Tucker MR, Leiser WL (2017) A modern green revolution gene for reduced height in wheat. Plant J. https://doi.org/10.1111/tpj.13726
Yu M, Mao SL, Chen GY, Pu ZE, Wei YM, Zheng YL (2014) QTLs for uppermost internode and spike length in two wheat RIL populations and their affect upon plant height at an individual QTL level. Euphytica 200:95e–108e. https://doi.org/10.1007/s10681-014-1156-7
Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, Korzun V, Argillier O, Stiewe G, Hinze M, Neumann K, Ganal MW, Roder MS (2014) Whole genome association mapping of plant height in winter wheat (Triticum aestivum L.). PLoS ONE. https://doi.org/10.1371/journal.pone.0113287
Zhai HJ, Feng ZY, Li J, Liu XY, Xiao SH, Ni ZF, Sun QX (2016) QTL analysis of spike morphological traits and plant height in winter wheat (Triticum aestivum L.) using a high-density SNP and SSR-based linkage map. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01617
Zhang XK, Yang SJ, Zhou Y, He ZH, Xia XC (2006) Distribution of the Rht-B1b, Rht-D1b and Rht8 reduced height genes in autumn-sown Chinese wheats detected by molecular markers. Euphytica 152:109e–116e. https://doi.org/10.1007/s10681-006-9184-6
Zhang L, Cheng Z, Qin R, Qiu Y, Wang JL, Cui X, Gu L, Zhang X, Guo X, Wang D, Jiang L, Wu CY, Wang H, Cao X, Wan J (2012) Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24:4407e–4421e. https://doi.org/10.1105/tpc.112.102269
Zhang JJ, Dell B, Biddulph B, Drake-Brockman F, Walker E, Khan N, Wong DB, Hayden M, Appels R (2013a) Wild-type alleles of Rht-B1 and Rht-D1 as independent determinants of thousand-grain weight and kernel number per spike in wheat. Mol Breed 32:771e–783e. https://doi.org/10.1007/s11032-013-9905-1
Zhang WP, Zhang L, Qiao LY, Wu J, Zhao GY, Jing RL, Lv WY, Jia JZ (2013b) Cloning and haplotype analysis of TaSTE, which is associated with plant height in bread wheat (Triticum aestivum L.). Mol Breed 31:47e–56e. https://doi.org/10.1007/s11032-012-9767-y
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (Grant No. 2017YFD0101004) and the National Natural Science Foundation of China (Grant No. 91435204). The authors would like to thank the IWGSC for pre-publication access to the RefSeq v1.0 sequence.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Yunbi Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chai, L., Chen, Z., Bian, R. et al. Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor Appl Genet 132, 1815–1831 (2019). https://doi.org/10.1007/s00122-019-03318-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03318-z