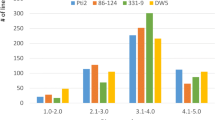
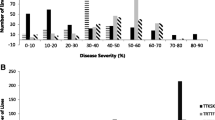
Abstract
Tan spot, caused by Pyrenophora tritici-repentis, is a major foliar disease of wheat worldwide. Host plant resistance is the best strategy to manage this disease. Traditionally, bi-parental mapping populations have been used to identify and map quantitative trait loci (QTL) affecting tan spot resistance in wheat. The association mapping (AM) could be an alternative approach to identify QTL based on linkage disequilibrium (LD) within a diverse germplasm set. In this study, we assessed resistance to P. tritici-repentis races 1 and 5 in 567 spring wheat landraces from the USDA-ARS National Small Grains Collection (NSGC). Using 832 diversity array technology (DArT) markers, QTL for resistance to P. tritici-repentis races 1 and 5 were identified. A linear model with principal components suggests that at least seven and three DArT markers were significantly associated with resistance to P. tritici-repentis races 1 and 5, respectively. The DArT markers associated with resistance to race 1 were detected on chromosomes 1D, 2A, 2B, 2D, 4A, 5B, and 7D and explained 1.3–3.1% of the phenotypic variance, while markers associated with resistance to race 5 were distributed on 2D, 6A and 7D, and explained 2.2–5.9% of the phenotypic variance. Some of the genomic regions identified in this study correspond to previously identified loci responsible for resistance to P. tritici-repentis, offering validation for our AM approach. Other regions identified were novel and could possess genes useful for resistance breeding. Some DArT markers associated with resistance to race 1 also were localized in the same regions of wheat chromosomes where QTL for resistance to yellow rust, leaf rust and powdery mildew, have been mapped previously. This study demonstrates that AM can be a useful approach to identify and map novel genomic regions involved in resistance to P. tritici-repentis.





Similar content being viewed by others
References
Abeysekara S, Friesen TL, Liu Z, McClean PE, Faris JD (2010) Marker development and saturation mapping of the tan spot Ptr ToxB sensitivity locus Tsc2 in hexaploid wheat. Plant Genome 3:179–189
Adhikari TB, Ali S, Myrfield M, Burlakoti RR (2008) The global genetic structure of Pyrenophora tritici-repentis populations. Phytopathology 98(Suppl.):S10
Agrama HA, Eizenga GC, Yan W (2007) Association mapping of yield and its components in rice cultivars. Mol Breeding 19:341–356
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113:1409–1420
Ali S, Francl LJ (2002) A new race of P. tritici-repentis from Brazil. (Abstract). Plant Dis 86:1050
Ali S, Gurung S, Adhikari TB (2010) Identification and characterization of novel isolates of Pyrenophora tritici-repentis from Arkansas. Plant Dis 94:229–235
Anderson JA, Effertz RJ, Faris JD, Francl LJ, Meinhardt SW, Gill BS (1999) Genetic analysis of sensitivity to a Pyrenophora tritici-repentis necrosis inducing toxin in durum and common wheat. Phytopathology 89:293–297
Arbelbide M, Bernardo R (2006) Mixed-model QTL mapping for kernel hardness and dough strength in bread wheat. Theor Appl Genet 112:885–890
Ballance GM, Lamari L, Bernier CC (1989) Purification and characterization of a host-selective necrosis toxin from Pyrenophora tritici-repentis. Physiol Mol Plant Pathol 35:203–213
Beattie AD, Edney MJ, Scoles GJ, Rossnagel BG (2010) Association mapping of malting quality data from western Canadian two-row barley cooperative trials. Crop Sci 50:1649–1663
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics 29:1165–1188
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664
Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177
Buckler ES IV, Thornsberry JM (2002) Plant molecular diversity and applications to genomics. Curr Opin Plant Biol 5:107–111
Chao S, Zhang W, Dubcovsky J, Sorrell M (2007) Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. Wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci 47:1018–1030
Chu CG, Friesen TL, Xu SS, Faris JD (2008) Identification of novel tan spot resistance loci beyond the known host-selective toxin insensitivity genes in wheat. Theor Appl Genet 117:873–881
Chu CG, Chao S, Friesen TL, Faris JD, Zhong SB, Xu SS (2010) Identification of novel tan spot resistance QTLs using an SSR-based linkage map of tetraploid wheat. Mol Breeding 25:327–338
Ciuffetti LM, Tuori RP (1999) Advances in the characterization of the Pyrenophora tritici-repentis-wheat interaction. Phytopathology 89:444–449
Ciuffetti LM, Tuori RP, Gaventa JM (1997) A single gene encodes a selective toxin causal to the development of tan spot of wheat. Plant Cell 9:135–144
Cook RJ, Yarham DJ (1989) Occurrence of tan spot of wheat caused by Pyrenophora tritici-repentis on wheat in England and Wales in 1987. Plant Pathol 38:101–102
Crossa J, Burgueno J, Dreisigacker S, Vargas M, Herrera Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177:1889–1913
de Wolf ED, Effertz RJ, Ali S, Francl L (1998) Vistas of tan spot research. Can J Plant Pathol 20:349–444
Effertz RJ, Anderson JA, Francl LJ (2001) Restriction fragment length polymorphism mapping of resistance to two races of Pyrenophora tritici-repentis in adult and seedling wheat. Phytopathology 91:572–578
Effertz RJ, Meinhardt SW, Anderson JA, Jordahl JG, Francl LJ (2002) Identification of a chlorosis-inducing toxin from Pyrenophora tritici-repentis and the chromosomal location of an insensitivity locus in wheat. Phytopathology 92:527–533
Elias E, Cantrell RG, Horsford RM Jr (1989) Heritability of resistance to tan spot in durum wheat and its association with other agronomic traits. Crop Sci 29:299–304
Emebiri LC, Oliver JR, Mrva K, Mares D (2010) Association mapping of late maturity α-amylase (LMA) activity in a collection of synthetic hexaploid wheat. Mol Breeding 26:39–49
Faris JD, Friesen TL (2005) Identification of quantitative trait loci for race-nonspecific resistance to tan spot in wheat. Theor Appl Genet 111:386–392
Faris JD, Anderson JA, Francl LJ, Jordahl JG (1996) Chromosomal location of a gene conditioning insensitivity in wheat to a necrosis-inducing culture filtrate from Pyrenophora tritici-repentis. Phytopathology 86:459–463
Faris JD, Anderson JA, Francl LJ, Jordhal JG (1997) RFLP mapping of resistance to chlorosis induction by Pyrenophora tritici-repentis in wheat. Theor Appl Genet 94:98–103
Faris JD, Li WL, Liu DJ, Chen PD, Gill BS (1999) Candidate gene analysis of quantitative disease resistance in wheat. Theor Appl Genet 98:219–225
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Friesen TL, Faris JD (2004) Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor Appl Genet 109:464–471
Gamba FM, Lamari L (1998) Mendelian inheritance of resistance to tan spot (Pyrenophora tritici-repentis) in selected genotypes of durum wheat (Triticum turgidum). Can J Plant Pathol 20:408–414
Gindrat D, Frei P, Mohl D (1988) The diagnostic and information service on diseases of major cultivated plants at the Changins station. Rev Suisse Agric 20:247–248
Goldstein DB, Tate SK, Sisodiya SM (2003) Pharmacogenetics goes genomic. Nat Rev Genet 4:937–947
Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38:226–231
Hosford RM Jr (1982) Tan spot-developing knowledge 1902–1981, virulent races and wheat differentials, methodology, rating systems, other leaf diseases, literature. In: Hosford RM Jr (ed) Tan spot of wheat and related diseases workshop. North Dakota Agricultural Experiment Station, Fargo, pp 1–24
Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S, McMullen MD, Ware D, Balint-Kurti PJ, Holland JB (2011) Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet 43:163–168
Lamari L, Bernier CC (1989) Evaluation of wheat lines and cultivars to tan spot (Pyrenophora tritici-repentis) based on lesion type. Can J Plant Pathol 11:49–56
Lamari L, Strelkov SE, Yahyaoui A, Orabi J, Smith RB (2003) The identification of two new races of Pyrenophora tritici- repentis from the host center of diversity confirms a one to one relationship in tan spot of wheat. Phytopathology 93:391–396
Li WL, Faris JD, Chittoor JM, Leach JE, Hulbert SH, Liu DJ, Chen PD, Gill BS (1999) Genomic mapping of defense response genes in wheat. Theor Appl Genet 98:226–233
Li HB, Yan W, Liu GR, Wen SM, Liu CJ (2011) Identification and validation of quantitative trait loci conferring tan spot resistance in the bread wheat variety Ernie. Theor Appl Genet 122:395–403
Liu K, Muse S (2004) PowerMarker: New Genetic Data Analysis Software, Version 2.7 (http://www.powermarker.net)
Maccaferri M, Sanguineti MC, Noli E, Tuberosa R (2005) Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Mol Breeding 15:271–289
Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA (2007) A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 75:879–889
Manning VA, Pandelova I, Ciuffetti LM (2002) A race for a novel host selective toxin. Phytopathology 92:S51
Martinez JP, Ottum SA, Ali S, Francl LJ, Ciuffetti LM (2001) Characterization of the ToxB gene from Pyrenophora tritici-repentis. Mol Plant-Microbe Interact 14:675–677
Massman J, Cooper B, Horsley R, Neate S, Macky RD, Chao S, Dong Y, Schwarz P, Muehlbauer GJ, Smith KP (2011) Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol Breeding 27:439–454
Meinhardt S, Ali S, Ling H, Francl L (2003) A new race of Pyrenophora tritici-repentis that produces a putative host-selective toxin. In: Rasmussen JB, Friesen TL, Ali S (eds) Proceedings of the fourth international wheat tan spot and spot blotch workshop. North Dakota Agric. Exp. Station, Fargo, pp 117–121
Neu CNS, Keller B (2002) Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45:737–744
Neumann K, Kobiljski B, Dencic S, Varshney RK, Borner A (2011) Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L.). Mol Breeding 27:37–58
Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ, Stahl EA, Weigel D (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet 30:190–193
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Raman H, Stodart B, Ryan PP, Delhaize E, Emebiri l, Raman R, Coombes N, Milgate A (2010) Genome-wide association analyses of common wheat (Triticum aestivum L.) germplasm identifies multiple loci for aluminium resistance. Genome 53:957–966
Roy JK, Smith KP, Muehlbauer GJ, Chao S, Close TJ, Steffenson BJ (2010) Association mapping of spot blotch resistance in wild barley. Mol Breeding 26:243–256
Sadeque A, Turner MA (2010) QTL analysis of plant height in hexaploid wheat doubled haploid population. Thai J Agric Sci 43(2):91–96
SAS Institute Inc. (2010) SAS OnlineDoc, Version 9.2. SAS Institute, Cary, USA
Scheet P, Stephens M (2006) A fast and flexible statistical model for large scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet 78:629–644
Schilder AMC (1989) Distribution, prevalence and severity of fungal leaf and spike disease of winter wheat in New York. Phytopathology 80:84–90
Shah DA, Madden LV (2004) Nonparametric analysis of ordinal data in designed factorial experiments. Phytopathology 94:33–43
Singh PK, Hughes GR (2006) Genetic similarity among isolates of Pyrenophora tritici-repentis, causal agent of tan spot of wheat. J Phytopathology 154:178–184
Singh PK, Gonzalez-Hernandez JL, Mergoum M, Ali S, Adhikari TB, Kianian SF, Elias EM, Hughes GR (2006) Identification and molecular mapping of a gene in tetraploid wheat conferring resistance to Pyrenophora tritici-repentis race 3. Phytopathology 96:885–889
Singh PK, Mergoum M, Gonzalez-Hernandez JL, Ali S, Adhikari TB, Kianian SF, Elias EM, Hughes GR (2008) Genetics and molecular mapping of resistance to necrosis inducing race 5 of Pyrenophora tritici-repentis in tetraploid wheat. Mol Breeding 21:293–304
Singh PK, Singh RP, Duveiller E, Mergoum M, Adhikari TB, Elias EM (2009) Genetics of wheat-Pyrenophora tritici-repentis interactions. Euphytica 171:1–13
Singh PK, Mergoum M, Adhikari TB, Shah T, Ghavami F, Kianian SF (2010) Genetic and molecular analysis of wheat tan spot resistance effective against Pyrenophora tritici-repentis races 2 and 5. Mol Breeding 25:369–379
Strelkov SE, Lamari L, Balance GM (1999) Characterization of a host-specifi c protein (Ptr ToxB) from Pyrenophora tritici-repentis. Mol Plant-Microbe Interact 12:728–732
Tadesse W, Hsam SLK, Zeller FJ (2006a) Evaluation of common wheat (Triticum aestivum L.) cultivars for tan spot resistance and chromosomal location of a resistance gene in cultivar ‘Salamouni’. Plant Breed 125:318–322
Tadesse W, Hsam SLK, Wenzel G, Zeller FJ (2006b) Identification and monosomic analysis of tan spot resistance genes in synthetic wheat lines (Triticum turgidum L. × Aegilops tauschii Coss.). Crop Sci 46:1212–1217
Tomas A, Feng GH, Reeck GR, Bockus WW, Leach JE (1990) Purification of a cultivar-specific toxin from Pyrenophora tritici-repentis, causal agent of tan spot of wheat. Mol Plant-Microbe Interact 3:221–224
Tommasini L, Schnurbusch T, Fossati D, Mascher F, Keller B (2007) Association mapping of Stagonospora nodorum blotch resistance in modern European winter wheat varieties. Theor Appl Genet 115:697–708
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Amer Statist Ass 58:236–244
Weber AL, Clark MR, Vaughn L, Sanchez-Gonzalez JDJ, Yu J, Yandell BS, Bradbury P, Doebley JF (2007) Major regulatory genes in maize contribute to standing variation in Teosinte (Zea mays ssp. parviglumis). Genetics 177:2349–2359
Weber AL, Briggs WH, Rucker J, Baltazar BM, de Jesus Sanchez-Gonzalez J, Feng P, Buckler E, Doebley JF (2008) The genetic architecture of complex traits in teosinte (Zea mays ssp. parviglumis): new evidence from association mapping. Genetics 180:1221–1232
Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotechnol 17:155–160
Acknowledgments
The authors gratefully acknowledge financial support for this project from the Wheat Research and Promotion Council, Minnesota, North Dakota Wheat Commission, and State Board of Agricultural Research and Education, North Dakota, and USDA-ARS specific cooperative agreement 58-5366-0-133. We are grateful to Dr. Andrzej Kilian for DArT marker data analysis and Jana Hansen for technical help.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Keller.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gurung, S., Mamidi, S., Bonman, J.M. et al. Identification of novel genomic regions associated with resistance to Pyrenophora tritici-repentis races 1 and 5 in spring wheat landraces using association analysis. Theor Appl Genet 123, 1029–1041 (2011). https://doi.org/10.1007/s00122-011-1645-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1645-1