Abstract
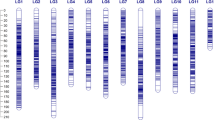
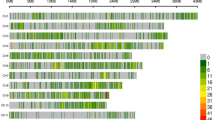
Drought tolerance in rice is controlled by several genes and is inherited quantitatively. Low genetic map density and the use of phenotypic traits that do not reflect the corresponding tolerance level have been obstacles in genetic analyses performed to identify genes that control drought-tolerant traits in rice. The current study aimed to construct a genetic map from high-density single-nucleotide polymorphism (SNP) markers generated from genome sequences of recombinant inbred lines (RILs), derived from IR64 × Hawara Bunar. Moreover, it sought to analyze the quantitative trait loci (QTL) and identify the drought tolerance candidate genes. A linkage map along 1980 cM on the 12 rice chromosomes was constructed employing 55,205 SNP markers resulting from the RIL genome sequences. A total of 175 morpho-physiological traits pertaining to drought stress were determined. A total of 41 QTLs were detected in 13 regions on rice chromosomes 1, 3, 6, 8, 9, and 12. Moreover, three hotspot QTL regions were found on chromosomes 6 and 8, along with two major QTL on chromosome 9. Differential gene expression for the loci within the QTL physical map intervals revealed many potential candidate genes. The markers tightly linked to the QTL and their candidate genes can potentially be used for pyramiding in marker-assisted breeding in order to achieve genetic improvement concerning the tolerance of rice to drought stress.





Similar content being viewed by others
References
Adams S, Grundy J, Veflingstad SR et al (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol 220:893–907
Alvarado G, Rodríguez FM, Pacheco A et al (2020) META-R: a software to analyze data from multi-environment plant breeding trials. Crop J 8:745–756
Anshori MF, Purwoko BS, Dewi IS et al (2019) Selection index based on multivariate analysis for selecting doubled-haploid rice lines in lowland saline prone area. SABRAO J Breed Genet 51:161–174
Ball JT, Woodrow IE, Berry JA (1987) A model predicting stomatal conductance and its contribution to the control of photosynthesis under different environmental conditions. Prog Photosynth Res 4:221–224
Bhattarai U, Subudhi PK (2018) Identification of drought responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP linkage map. Euphytica 214:1–17
Bidinger FR, Mahalakshmi V, Rao GDP (1987) Assessment of drought resistance in pearl millet (Pennisetum americanum (L.) Leeke). Aust J Agric Res 38:37–48
Borah P, Sharma E, Kaur A et al (2017) Analysis of drought-responsive signalling network in two contrasting rice cultivars using transcriptome-based approach. Sci Rep 7:1–21
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Cao X, Zhu C, Zhong C et al (2018) Mixed-nitrogen nutrition-mediated enhancement of drought tolerance of rice seedlings associated with photosynthesis, hormone balance and carbohydrate partitioning. Plant Growth Regul 84:451–465
Chen GB, Zhu ZX, Zhang FT, Zhu J (2012) Quantitative genetic analysis station for the genetic analysis of complex traits. Chin Sci Bull 57:2721–2726
Chen G, Zou Y, Hu J, Ding Y (2018) Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics 19:1–14
Cui D, Wu D, Somarathna Y et al (2015) QTL mapping for salt tolerance based on snp markers at the seedling stage in maize (Zea mays L.). Euphytica 203:273–283
Curtis TY, Bo V, Tucker A, Halford NG (2018) Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress. Food Energy Secur 7:1–16
Daniel B, Pavkov-Keller T, Steiner B et al (2015) Oxidation of monolignols by members of the berberine bridge enzyme family suggests a role in plant cell wall metabolism. J Biol Chem 290:18770–18781
Dashti H, Yazdi-Samadi B, Ghannadha M et al (2007) QTL analysis for drought resistance in wheat using doubled haploid lines. Int J Agric Biol 9:98–102
Davar R, Darvishzadeh R, Majd A et al (2020) QTL mapping of partial resistance to basal stem rot in sunflower using recombinant inbred lines. Phytopathol Mediterr 49:330–341
De Leon TB, Linscombe S, Subudhi PK (2016) Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 9:1–22
Dobin A, Davis CA, Schlesinger F et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6:1–10
Fan X, Cui F, Ji J et al (2019) Dissection of pleiotropic QTL regions controlling wheat spike characteristics under different nitrogen treatments using traditional and conditional QTL mapping. Front Plant Sci 10:1–13
Fendiyanto MH, Satrio RD, Suharsono S et al (2019a) QTL for aluminum tolerance on rice chromosome 3 based on root length characters. SABRAO J Breed Genet 51:451–469
Fendiyanto MH, Satrio RD, Suharsono S et al (2019b) Correlation among Snpb11markers, root growth, and physiological characters of upland riceunder aluminum stress. Biodiversitas 20:1243–1254
Fischer RA, Maurer R (1978) Drought resistance in spring wheat cultivars, I: grain yield responses. Aust J Agric Res 29:897–912
Fischer RA, Wood JT (1979) Drought resistance in spring wheat cultivars, III: yield associations with morpho-physiological traits. Aust J Agric Res 30:1001–1020
Fukino N, Yoshioka Y, Sugiyama M et al (2013) Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol Bred 32:267–277
Funatsuki H, Kawaguchi K, Matsuba S et al (2005) Mapping of QTL associated with chilling tolerance during reproductive growth in soybean. Theor Appl Genet 111:851–861
Giuliani R, Koteyeva N, Voznesenskaya E et al (2013) Coordination of leaf photosynthesis, transpiration, and structural traits in rice and wild relatives (Genus Oryza). Plant Physiol 162:1632–1651
Harushima Y, Yano M, Shomura A et al (1998) A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148:479–494
Healey A, Furtado A, Cooper T, Henry RJ (2014) Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10:1–8
Hemamalini GS, Shashidhar HE, Hittalmani S (2000) Molecular marker assisted tagging of morphological and physiological traits under two contrasting moisture regimes at peak vegetative stage in rice (Oryza sativa L.). Euphytica 112:69–78
Jiang SC, Mei C, Liang S et al (2015) Crucial roles of the pentatricopeptide repeat protein SOAR1 in Arabidopsis response to drought, salt and cold stresses. Plant Mol Biol 88:369–385
Jing Y, Lin R (2015) The VQ motif-containing protein family of plant-specific transcriptional regulators. Plant Physiol 169:371–378
Jung YJ, Melencion SMB, Lee ES et al (2015) Universal stress protein exhibits a redox-dependent chaperone function in arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front Plant Sci 6:1–11
Kadam NN, Yin X, Bindraban PS et al (2015) Does morphological and anatomical plasticity during the vegetative stage make wheat more tolerant of water deficit stress than rice? Plant Physiol 167:1389–1401
Kalladan R, Worch S, Rolletschek H et al (2013) Identification of quantitative trait loci contributing to yield and seed quality parameters under terminal drought in barley advanced backcross lines. Mol Breed 32:71–90
Kamies R, Farrant J, Tadele Z et al (2017) A Proteomic approach to investigate the drought response in the orphan crop Eragrostis tef. Proteomes 5:32
Kaminski KP, Kørup K, Kristensen K et al (2015) Contrasting water-use efficiency (WUE) responses of a potato mapping population and capability of modified ball-berry model to predict stomatal conductance and WUE measured at different environmental conditions. J Agron Crop Sci 201:81–94
Kawahara Y, de la Bastide M, Hamilton JP et al (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:1–10
Kimbara J, Ohyama A, Chikano H et al (2018) QTL mapping of fruit nutritional and flavor components in tomato (Solanum lycopersicum) using genome-wide SSR markers and recombinant inbred lines (RILs) from an intra-specific cross. Euphytica 214:210
Kinsella RJ, Kähäri A, Haider S et al (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011:1–9
Kumar R, Subba A, Kaur C et al (2017) OsCBSCBSPB4 is a two cystathionine-β-synthase domain-containing protein from rice that functions in abiotic stress tolerance. Curr Genomics 19:50–59
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760
Li Z, Mu P, Li C et al (2005) QTL mapping of root traits in a doubled haploid population from a cross between upland and lowland japonica rice in three environments. Theor Appl Genet 110:1244–1252
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Liu Z, Yan JP, Li DK et al (2015) UDP-glucosyltransferase71C5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol 167:1659–1670
Liu WC, Li YH, Yuan HM et al (2017) WD40-REPEAT 5a functions in drought stress tolerance by regulating nitric oxide accumulation in Arabidopsis. Plant Cell Environ 40:543–552
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21
Ma C, Burd S, Lers A (2015) MiR408 is involved in abiotic stress responses in Arabidopsis. Plant J 84:169–187
Mason RE, Mondal S, Beecher FW et al (2010) QTL associated with heat susceptibility index in wheat (Triticum aestivum L.) under short-term reproductive stage heat stress. Euphytica 174:423–436
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Miftahudin M, Putri RE, Chikmawati T (2020) Vegetative morphophysiological responses of four rice cultivars to drought stress. Biodiversitas 21:3727–3734
Miftahudin M, Roslim DI, Fendiyanto MH, Satrio RD et al (2021) OsGERLP: A novel aluminum tolerance rice gene isolated from a local cultivar in Indonesia. Plant Physiol Biochem 162:86–99
Moncada P, Martínez CP, Borrero J et al (2001) Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor Appl Genet 102:41–52
Moosavi SS, Samadi YB, Dashti H, Pourshahbazi A (2008) Introduction of new indices to identify relative drought tolerance and resistance in wheat genotypes. Desert 5:165–178
Nishanth MJ, Sheshadri SA, Rathore SS et al (2018) Expression analysis of cell wall invertase under abiotic stress conditions influencing specialized metabolism in Catharanthus roseus. Sci Rep 8:1–15
Oladosu Y, Rafii MY, Samuel C et al (2019) Drought resistance in rice from conventional to molecular breeding. Int J Mol Sci 20:3519
Pandey V, Shukla A (2015) Acclimation and tolerance strategies of rice under drought stress. Rice Sci 22:147–161
Peterson RA, Cavanaugh JE (2019) Ordered quantile normalization: a semiparametric transformation built for the cross-validation era. J Appl Stat 0:1–16
Price AH, Townend J, Jones MP, Audebert A (2002) Mapping QTLs associated with drought avoidance in upland rice grown in the Philippines and West Africa. Plant Mol Biol 48:683–695
Prince SJ, Beena R, Gomez SM et al (2015) Mapping consistent rice (Oryza sativa L.) yield QTLs under drought stress in target rainfed environments. Rice 8:1–13
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Rao DE, Chaitanya KV (2016) Photosynthesis and antioxidative defense mechanisms in deciphering drought stress tolerance of crop plants. Biol Plant 60:201–218
Rehman HM, Nawaz MA, Shah ZH et al (2018) Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci Rep 8:1–18
Rosielle AA, Hamblin J (1981) Theoretical aspects of selection for yield in stress and non-stress environments. Crop J 21:934–946
Rueden CT, Schindelin J, Hiner MC et al (2017) Image J2: ImageJ for the next generation of scientific image data. BMC Bioinformat 18:1–26
Sabar M, Shabir G, Shah SM et al (2019) Identification and mapping of QTLs associated with drought tolerance traits in rice by a cross between super Basmati and IR55419-04. Breed Sci 69:169–178
Satrio RD, Fendiyanto MH, Supena EDJ et al (2019) Identification of drought-responsive regulatory genes by hierarchical selection of expressed sequence tags and their expression under drought stress in rice. Intl J Agric Biol 22:1524–1532
Spindel J, Wright M, Chen C et al (2013) Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet 126:2699–2716
Takahashi F, Kuromori T, Sato H, Shinozaki K (2018) Regulatory gene networks in drought stress responses and resistance in plants. In: Iwaya-Inoue M, Sakurai M, Uemura M (eds) Survival strategies in extreme cold and desiccation, advances in experimental medicine and biology. Springer, Singapore, pp 189–214
Tanyolac B, Ozatay S, Kahraman A, Muehlbauer F (2010) Linkage mapping of lentil (Lens culinaris L.) genome using recombinant inbred lines revealed by AFLP, ISSR, RAPD and some morphologic markers. J Agric Biotechnol Sustain Dev 2:1–6
Todaka D, Takahashi F, Yamaguchi-Shinozaki K, Shinozaki K (2019) ABA-responsive gene expression in response to drought stress: cellular regulation and long-distance signaling. Elsevier, Amsterdam
Uga Y, Okuno K, Yano M (2011) Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J Exp Bot 62:2485–2494
Vijayalakshmi K, Fritz AK, Paulsen GM et al (2012) Modeling and mapping QTL for senescence-related traits in winter wheat under high temperature. Mol Bred 26:163–175
Vinod KK, Krishnan SG, Thribhuvan R, Singh AK (2019) Genetics of drought tolerance, mapping QTLs, candidate genes and their utilization in rice improvement. In: Rajpal V, Sehgal D, Kumar A, Raina S (eds) Genomics assisted breeding of crops for abiotic stress tolerance. Springer, Cham, pp 145–186
Wang X, Jiang G, Green M, Scott RA, Hyten DL, Cregan PB (2012) Quantitative trait locus analysis of saturated fatty acids in a population of recombinant inbred lines of soybean. Mol Bred 30:1163–1179
Xu Y, Wang R, Tong Y et al (2014) Mapping QTLs for yield and nitrogen-related traits in wheat: influence of nitrogen and phosphorus fertilization on QTL expression. Theor Appl Genet 127:59–72
Yao W, Li G, Cui Y et al (2019) Mapping quantitative trait loci using binned genotypes. J Genet Genomics 46:343–352
Yue B, Xiong L, Xue W et al (2005) Genetic analysis for drought resistance of rice at reproductive stage in field with different types of soil. Theor Appl Genet 111:1127–1136
Zhang J, Zheng HG, Aarti A et al (2001) Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor Appl Genet 103:19–29
Zhang H, Cui F, Wang L et al (2013) Conditional and unconditional QTL mapping of drought-tolerance-related traits of wheat seedling using two related RIL populations. J Genet 92:213–231
Zhou L, Liu Y, Liu Z et al (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61:4157–4168
Zhu J (1995) Analysis of conditional genetic effects and variance components in developmental genetics. Genetics 141:1633–1639
Zu X, Lu Y, Wang Q et al (2017) A new method for evaluating the drought tolerance of upland rice cultivars. Crop J 5:488–498
Fernandez GCJ (1992) Stress tolerance index - a new indicator of tolerance. Horticult Sci 27:626–626.
Acknowledgements
This work was supported by funding from the Ministry of Research and Technology, Indonesia. We thank Prof. Taku Demura, Dr. Tadashi Kunieda, and Dr. Ryosuke Sano from Nara Institute of Science and Technology (NAIST), Japan, for assisting us in conducting genotyping-by-sequencing analysis.
Funding
This work was funded by the World Class Research (WCR) F.Y. 2019–2020, Grant No. 1536/IT3.L1/PN/2019, awarded to Dr. Miftahudin from the Ministry of Research and Technology, the Republic of Indonesia. The research was also partially supported by the ‘Program Magister Menuju Doktor untuk Sarjana Unggul’ (PMDSU) F.Y. 2016–2019 from the Ministry of Research, Technology, and Higher Education, the Republic of Indonesia.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. The population of recombinant inbred lines was developed by MM. RDS phenotyped and analyzed the RIL data, performed DNA isolation, constructed the high-resolution genetic map, ran the quantitative trait loci analysis of the RILs, and wrote the manuscript under the supervision of EDJS, SS, and MM. MHF performed DNA isolation and phenotyped the RILs. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Satrio, R.D., Fendiyanto, M.H., Supena, E.D.J. et al. Genome-wide SNP discovery, linkage mapping, and analysis of QTL for morpho-physiological traits in rice during vegetative stage under drought stress. Physiol Mol Biol Plants 27, 2635–2650 (2021). https://doi.org/10.1007/s12298-021-01095-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01095-y