Abstract
Background
In rice, drought stress at reproductive stage drastically reduces yield, which in turn hampers farmer’s efforts towards crop production. The majority of the rice varieties have resistance genes against several abiotic and biotic stresses. Therefore, the traditional landraces were studied to identify QTLs/candidate genes associated with drought tolerance.
Methods and results
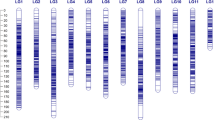
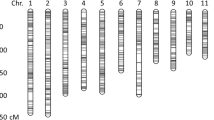
A high-density SNP-based genetic map was constructed using a Genotyping-by-sequencing (GBS) approach. The recombinant inbred lines (RILs) derived from crossing ‘Banglami × Ranjit’ were used for QTL analysis. A total map length of 1306.424 cM was constructed, which had an average inter-marker distance of 0.281 cM. The phenotypic evaluation of F6 and F7 RILs were performed under drought stress and control conditions. A total of 42 QTLs were identified under drought stress and control conditions for yield component traits explaining 1.95–13.36% of the total phenotypic variance (PVE). Among these, 19 QTLs were identified under drought stress conditions, whereas 23 QTLs were located under control conditions. A total of 4 QTLs explained a PVE ≥ 10% which are considered as the major QTLs. Moreover, bioinformatics analysis revealed the presence of 6 candidate genes, which showed differential expression under drought and control conditions.
Conclusion
These QTLs/genes may be deployed for marker-assisted pyramiding to improve drought tolerance in the existing rice varieties.


Similar content being viewed by others
References
Nachimuthu VV, Sabariappan R, Muthurajan R, Kumar A (2017) Breeding rice varieties for abiotic stress tolerance: challenges and opportunities. Abiotic stress management for resilient agriculture. Springer, Singapore, pp 339–361
Verma RK, Chetia SK, Dey PC, Sen P, Modi MK (2017) Breeding for drought tolerance–a major challenge for rice cultivation under water limiting conditions. J Pharm Phytochem 7(5):813–816
Chetia SK, Kalita M, Verma RK, Barua B, Ahmed T, Modi MK, Singh NK (2018) Flood proofing of popular North-Eastern India rice variety Ranjit by simplified marker-assisted backcross breeding of Sub1 gene. Indian J Genet 78(2):166–173
Yang X, Wang B, Chen L, Li P, Cao C (2019) The different influences of drought stress at the flowering stage on rice physiological traits grain yield and quality. Sci Rep 9(1):1–12
Verma RK, Chetia SK, Tamuly A, Sharma V, Dey PC, Sen P, Modi MK (2021) Characterization of winter rice (Oryza sativa L.) germplasm of North East India using morphological traits. Indian J Trad Knowl 20(3):838–845
Arora A, Bansal S, Ward PS (2019) Do farmers value rice varieties tolerant to droughts and floods? Evidence from a discrete choice experiment in Odisha India. Water Resourc Econ 25:27–41
Verma RK, Chetia SK, Sharma V, Devi K, Kumar A, Modi MK (2022) Identification and characterization of genes for drought tolerance in upland rice cultivar ‘Banglami’ of North East India. Mol Biol Rep. https://doi.org/10.1007/s11033-022-07859-3
Neog P, Sarma PK, Saikia D (2020) Management of drought in sali rice under increasing rainfall variability in the north bank plains zone of Assam, North East India. Clim Change 158(3):473–484
Verma RK, Chetia SK, Dey PC, Baruah AR, Modi MK (2017) Mapping of QTLs for grain yield and its component traits under drought stress in elite rice variety of Assam. Int J Curr Microbiol App Sci 6(6):1443–1455
Sandhu N, Kumar A (2017) Bridging the rice yield gaps under drought: QTLs genes and their use in breeding programs. Agronomy 7(2):27
Verma RK, Chetia SK, Dey PC, Rahman A, Saikia S, Sharma V, Sharma H et al (2021) Genome-wide association studies for agronomical traits in winter rice accessions of Assam. Genomics 113(3):1037–1047
Verma RK, Chetia SK, Sharma V, Baishya S, Sharma H, Modi MK (2022) GWAS to spot candidate genes associated with grain quality traits in diverse rice accessions of North East India. Mol Biol Rep 49:5365–5377
Rasheed A, Hao Y, Xia X, Khan A, Xu Y, Varshney RK, He Z (2017) Crop breeding chips and genotyping platforms: progress challenges and perspectives. Mol Plant 10(8):1047–1064
Yadav S, Sandhu N, Singh VK, Catolos M, Kumar A (2019) Genotyping-by-sequencing based QTL mapping for rice grain yield under reproductive stage drought stress tolerance. Sci Rep 9(1):1–12
Levy SE, Boone BE (2019) Next-generation sequencing strategies. Cold Spring Harbor Perspec Med 9(7):a025791
Mishra A, Singh PK, Bhandawat A, Sharma V, Sharma V, Singh P, Roy J et al (2022) Analysis of SSR and SNP markers. Bioinformatics. Academic Press, Cambridge, pp 131–144
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):e19379
De Leon TB, Linscombe S, Subudhi PK (2016) Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice 9(1):52
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T et al (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform 43(1):11–20
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE et al (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3(3):269–283
Li H, Hearne S, Bänziger M, Li Z, Wang J (2010) Statistical properties of QTL linkage mapping in biparental genetic populations. Heredity 105(3):257–267
Wang J (2009) Inclusive composite interval mapping of quantitative trait genes. Acta Agron Sin 35(2):239–245
McCouch SR (2008) Gene nomenclature system for rice. Rice 1(1):72–84
Kulkarni SR, Balachandran SM, Ulaganathan K, Balakrishnan D, Prasad AS, Rekha G, Kousik MB et al (2021) Mapping novel QTLs for yield related traits from a popular rice hybrid KRH-2 derived doubled haploid (DH) population. 3 Biotech 11(12):1–20
Panda D, Mishra SS, Behera PK (2021) Drought tolerance in rice: Focus on recent mechanisms and approaches. Rice Sci 28(2):119–132
Liu JX, Liao DQ, Oane R, Estenor L, Yang XE, Li ZC, Bennett J (2006) Genetic variation in the sensitivity of anther dehiscence to drought stress in rice. Field Crop Res 97(1):87–100
Suji KK, Biji KR, Poornima R, Prince K, Amudha K, Kavitha S, Mankar S et al (2012) Mapping QTLs for plant phenology and production traits using indica rice (Oryza sativa L.) lines adapted to rainfed environment. Mol Biotechnol 52(2):151–60
Li FW, Harkess A (2018) A guide to sequence your favorite plant genomes. Appl in Plant Sci 6(3):e1030
Scheben A, Batley J, Edwards D (2018) Revolution in genotyping platforms for crop improvement. Plant genetics molecular biology. . Springer, Cham, pp 37–52
Sharma V, Verma RK, Dey PC, Chetia SK, Baruah AR, Modi MK (2017) QTLs associated with yield attributing traits under drought stress in upland rice cultivar of Assam. Oryza 54:253–257
Satrio RD, Fendiyanto MH, Supena ED, Suharsono S, Miftahudin M (2021) Genome-wide SNP discovery, linkage mapping, and analysis of QTL for morpho-physiological traits in rice during vegetative stage under drought stress. Physiol Mol Biol Plants 27(11):2635–2650
Mahalle MD, Dey PC, Chetia SK (2021) Association mapping for yield traits under drought stress in Autumn rice germplasm collection of Assam. J Plant Biochem Biotech 30(1):26–36
Bernier J, Kumar A, Ramaiah V, Spaner D, Atlin G (2007) A large-effect QTL for grain yield under reproductive-stage drought stress in upland rice. Crop Sci 47(2):507–516
Verma RK, Chetia SK, Dey PC, Sharma V, Baruah AR, Modi MK (2017) Development of advanced breeding lines for high grain yield under drought stress in elite rice genetic background. Crop Res 18(4):713–718
Hussien A, Tavakol E, Horner DS, Muñoz-Amatriaín M, Muehlbauer GJ, Rossini L (2014) Genetics of tillering in rice and barley. Plant Genom 7(1):2013–2010
Verma RK, Dey PC, Chetia SK, Modi MK (2017) Development of Advanced breeding lines for drought tolerance based on physiological and yield traits. Oryza 54(2):169–173
Barik SR, Pandit E, Mohanty SP, Nayak DK, Pradhan SK (2020) Genetic mapping of physiological traits associated with terminal stage drought tolerance in rice. BMC Genet 21:76
Barik SR, Pandit E, Pradhan SK, Mohanty SP, Mohapatra T (2019) Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE 14(12):e0214979
Faraji S, Filiz E, Kazemitabar SK, Vannozzi A, Palumbo F, Barcaccia G, Heidari P (2020) The AP2/ERF gene family in Triticum durum: genome-wide identification and expression analysis under drought and salinity stresses. Genes 11(12):1464
Yang Y, Wang W, Chu Z, Zhu JK, Zhang H (2017) Roles of nuclear pores and nucleo-cytoplasmic trafficking in plant stress responses. Front Plant Sci 8:574
Jia Q, Kong D, Li Q, Sun S, Song J, Zhu Y, Liang K et al (2019) The function of inositol phosphatases in plant tolerance to abiotic stress. IJMS 20(16):3999
Pandian BA, Sathishraj R, Djanaguiraman M, Prasad PV, Jugulam M (2020) Role of cytochrome P450 enzymes in plant stress response. Antioxidants 9(5):454
Ueda M, Seki M (2020) Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol 182(1):15–26
Banerjee A, Roychoudhury A (2018) The gymnastics of epigenomics in rice. Plant Cell Rep 37(1):25–49
Liang X, Zhou JM (2018) Receptor-like cytoplasmic kinases: central players in plant receptor kinase–mediated signaling. Ann Rev Plant Biol 69:267–299
Acknowledgements
The authors are highly thankful to DBT-NECAB, AAU Jorhat and Department of Agricultural Biotechnology (ABT), AAU, Jorhat for financial and technical help during the entire research programme. The authors are also grateful towards the contribution of NextGen BioSciences Diagnostics Private Limited, New Delhi for sequencing purposes. The authors also acknowledge Distributed Information Centre (DIC), Department of ABT, AAU, Jorhat for providing necessary help during GBS data analysis. The authors also appreciate the help of Dr. Anjan Gowda S. North Carolina State University, Raleigh, North Carolina, United States for his immense support during the data analysis and critical explanations on different parts of the research work.
Author information
Authors and Affiliations
Contributions
Conceptualization: (MKM); Investigation: (NR, RKV, PS, MKM); Resources: (SKC, MKM, RKV); Methodology: (RKV, NR, VS, MKM); Data analysis (NR, RKV); Writing, review and editing (NR, RKV, VS, MKM).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The current study does not involve any animal or human study.
Informed consent
Not applicable for current study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Article has been submitted to a preprint platform with https://doi.org/10.21203/rs.3.rs-1813071/v1. This work is licensed under a CC BY 4.0 License.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Roy, N., Verma, R.K., Chetia, S.K. et al. Molecular mapping of drought-responsive QTLs during the reproductive stage of rice using a GBS (genotyping-by-sequencing) based SNP linkage map. Mol Biol Rep 50, 65–76 (2023). https://doi.org/10.1007/s11033-022-08002-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08002-y