Abstract
Background
Drought stress is a major constraint for rice production worldwide. Reproductive stage drought stress (RSDS) leads to heavy yield losses in rice. The prospecting of new donor cultivars for identification and introgression of QTLs of major effect (Quantitative trait locus) for drought tolerance is crucial for the development of drought-resilient rice varieties.
Methods and results
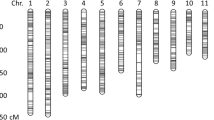
Our study aimed to map QTLs associated with yield and its related traits under RSDS conditions. A saturated linkage map was constructed using 3417 GBS (Genotyping by sequencing) derived SNP (Single nucleotide polymorphism) markers spanning 1924.136 cM map length with an average marker density of 0.56 cM, in the F3 mapping population raised via cross made between the traditional ahu rice cultivar, Koniahu (drought tolerant) and a high-yielding variety, Disang (drought susceptible). Using the Inclusive composite interval mapping approach, 35 genomic regions governing yield and related traits were identified in pooled data from 198 F3 and F4 segregating lines evaluated for two consecutive seasons under both RSDS and irrigated control conditions. Of the 35 QTLs, 23 QTLs were identified under RSDS with LOD (Logarithm of odds) values ranging between 2.50 and 7.83 and PVE (phenotypic variance explained) values of 2.95–12.42%. Two major QTLs were found to be linked to plant height (qPH1.29) and number of filled grains per panicle (qNOG5.12) under RSDS. Five putative QTLs for grain yield namely, qGY2.00, qGY5.05, qGY6.16, qGY9.19, and qGY10.20 were identified within drought conditions. Fourteen QTL regions having ≤ 10 Mb QTL interval size were further analysed for candidate gene identification and a total of 4146 genes were detected out of these 2263 (54.63%) genes were annotated to at least one gene ontology (GO) term.
Conclusion
Several QTLs associated with grain yield and yield components and putative candidate genes were identified. The putative QTLs and candidate genes identified could be employed to augment drought resilience in rice after further validation through MAS strategies.



Similar content being viewed by others
References
Kurata N, Nonomura K, Harushima Y (2002) Rice genome organization: the centromere and genome interactions. Ann Bot 90(4):427–435. https://doi.org/10.1093/aob/mcf218
Srinivas T (2011) Exploring indian culture through food. Food Cult Asia 16(3):38–41
FAO (2020) Food and Agriculture Organization of the United Nations. FAOSTAT statistical database
Singh R, SinghY, Xalaxo S et al (2016) From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci 242:278–287. https://doi.org/10.1016/j.plantsci.2015.08.008
Dar MH, Waza SA, Shukla S, Zaidi NW, Nayak S, Hossain M, Kumar A, Ismail AM, Singh US (2020) Drought tolerant rice for ensuring food security in eastern India. Sustainability 12(6):2214. https://doi.org/10.3390/su12062214
Pandey S, Bhandari H (2009) Drought: economic costs and research implications. In: Pandey S, Bhandari H, Hardy B (eds) Drought frontiers in rice: crop improvement for increased rainfed production. World Scientific, pp 3–17. https://doi.org/10.1142/9789814280013_0001
Parida BR, Oinam B (2015) Unprecedented drought in North East India compared to western India. Curr Sci 109(11):2121–2126. https://www.jstor.org/stable/24906713
Kumar A, Dixit S, Ram T, Yadaw RB, Mishra KK, Mandal NP (2014) Breeding high-yielding drought-tolerant rice: genetic variations and conventional and molecular approaches. J Exp Bot 65(21):6265–6278. https://doi.org/10.1093/jxb/eru363
Kumar A, Sandhu N, Dixit S, Yadav S, Swamy BPM, Shamsudin NAA (2018) Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 11(1):35. https://doi.org/10.1186/s12284-018-0227-0
Sandhu N, Dixit S, Swamy BPM et al (2019) Marker assisted breeding to develop multiple stress tolerant varieties for Flood and Drought Prone Areas. Rice 12(1):8. https://doi.org/10.1186/s12284-019-0269-y
Hore DK (2005) Rice diversity collection, conservation and management in northeastern India. Genet Resour Crop Evol 52(8):1129–1140. https://doi.org/10.1007/s10722-004-6084-2
Verma RK, Dey PC, Chetia SK, Modi MK (2017) Development of advanced breeding lines for drought tolerance based on physiological and yield traits. Oryza 54(2):169–173. https://doi.org/10.5958/2454-1761.2017.00015.8
Verma RK, Chetia SK, Tamuly A, Sharma V, Dey PC, Sen P, Modi MK (2021) Characterization of winter rice (Oryza sativa L.) germplasm of North East India using morphological traits. Indian J of Traditional Knowledge 20(3):838–845
Mahalle MD, Dey PC, Chetia SK, Baruah AR, Ahmed T, Sarma RN, Kaldate RC, Kumar A, Singh SK, Modi MK (2020) Association mapping for yield traits under drought stress in Autumn rice germplasm collection of Assam. J Plant Biochem Biotechnol 30(1):26–36. https://doi.org/10.1007/s13562-020-00559-8
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):e19379. https://doi.org/10.1371/journal.pone.0019379
Rasheed A, Hao Y, Xia X, Khan A, Xu Y, Varshney RK, He Z (2017) Crop breeding chips and genotyping platforms: Progress, Challenges, and perspectives. Mol Plant 10(8):1047–1064. https://doi.org/10.1016/j.molp.2017.06.008
Yadav S, Sandhu N, Singh VK, Catolos M, Kumar A (2019) Genotyping-by-sequencing based QTL mapping for rice grain yield under reproductive stage drought stress tolerance. Sci Rep 9(1):14326. https://doi.org/10.1038/s41598-019-50880-z
Kulkarni SR, Balachandran SM, Ulaganathan K et al (2020) Molecular mapping of QTLs for yield related traits in recombinant inbred line (RIL) population derived from the popular rice hybrid KRH-2 and their validation through SNP genotyping. Sci Rep 10(1):13695. https://doi.org/10.1038/s41598-020-70637-3
Verma RK, Chetia SK, Baishya S, Sharma V, Sharma H, Modi MK (2022) GWAS to spot candidate genes associated with grain quality traits in diverse rice accessions of North East India. Mol Biol Rep. https://doi.org/10.1007/s11033-021-07113-2
Verma RK, Chetia SK, Dey PC, Rahman A, Saikia S, Sharma V, Sharma H, Sen P, Modi MK (2020) Genome-wide association studies for agronomical traits in winter rice accessions of Assam. Genomics 113(3):1037–1047. https://doi.org/10.1016/j.ygeno.2020.11.033
Mahalle MD, Chetia SK, Dey PC, Sarma RN, Baruah AR, Kaldate RC, Verma RK, Modi MK (2022) Assessing the leaf shape dynamic through marker–trait association under drought stress in a rice germplasm panel. Plant Genet Resour 19(6):477–483. https://doi.org/10.1017/S1479262121000587
Chowdhury P, Pathak PK, Tripathy AK, Neog M, Saud RK, Saharia RR, Bharali M, Nath D, Saikia N, Borah D, Rahman A, Saikia S, Sarkar L, Biswas S (2019) Disang- a promising short duration rice variety suitable as early ahu (pre flood) and for post flood situation in Cachar district of Assam. Int J Agric Crop Sci 11(15):8881–8882
Federer W (1956) Augmented (or hoonuiaku) designs hawaiian Planters Record. In): Honolulu
IBMCorp Released 2012. IBM SPSS Statistics for Windows, Version 21.0. Armonk, NY: IBM Corp
Kumar A, Sharma D, Tiwari A, Jaiswal JP, Singh NK, Sood S (2016) Genotyping-by-sequencing analysis for determining population structure of finger millet germplasm of diverse origins. Plant Genome 9(2). https://doi.org/10.3835/plantgenome2015.07.0058
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Van Ooijen J (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res 93(5):343–349. https://doi.org/10.1017/S0016672311000279
Kosambi D (1944) The estimation of map distance from recombination values. Annals of Eugenics 12(1):172–175
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3(3):269–283. https://doi.org/10.1016/j.cj.2015.01.001
Bhattarai U, Subudhi PK (2018) Identification of drought-responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP. Link map Euphytica 214(2):38. https://doi.org/10.1007/s10681-018-2117-3
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78. https://doi.org/10.1093/jhered/93.1.77
Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2. 0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1):W122–W129. https://doi.org/10.1093/nar/gkx382
Bhattarai U, Subudhi PK (2018) Identification of drought-responsive QTLs during vegetative growth stage of rice using a saturated GBS-based SNP. Link map Euphytica 214(2):38. https://doi.org/10.1007/s10681-018-2117-3
Saikumar S, Saiharini A, Ayyappa D, Padmavathi G, Shenoy VV (2014) Heritability, correlation and path analysis among yield and yield attributing traits for drought tolerance in an interspecific cross derived from oryza sativa x o. glaberrima introgression line under contrasting moisture regimes. Notulae Scientia Biologicae 6(3):338–348. https://doi.org/10.15835/nsb639402
Barik SR, Pandit E, Pradhan SK, Mohanty SP, Mohapatra T (2019) Genetic mapping of morpho-physiological traits involved during reproductive stage drought tolerance in rice. PLoS ONE 14(12):e0214979. https://doi.org/10.1371/journal.pone.0214979
Prince SJ, Beena R, Gomez SM, Senthivel S, Babu RC (2015) Mapping consistent Rice (Oryza sativa L.) yield QTLs under drought stress in target rainfed environments. Rice 8(1):53. https://doi.org/10.1186/s12284-015-0053-6
Uddin MN, Tomita A, Obara M, Yanagihara S, Fukuta Y (2016) Identification of a low tiller gene from a new plant type cultivar in rice (Oryza sativa L). Breed Sci 66(5):790–796. https://doi.org/10.1270/jsbbs.16143
Kamoshita A, Babu RC, Boopathi NM, Fukai S (2008) Phenotypic and genotypic analysis of drought-resistance traits for development of rice cultivars adapted to rainfed environments. Field Crops Res 109(1–3):1–23. https://doi.org/10.1016/j.fcr.2008.06.010
Roy N, Verma RK, Chetia SK, Sharma V, Sen P, Modi MK (2022) Molecular mapping of drought-responsive QTLs during the reproductive stage of rice using a GBS (genotyping-by-sequencing) based SNP linkage map. Mol Biol Rep 50, 65–76 (2023). https://doi.org/10.1007/s11033-022-08002-y
Paran I, Goldman I, Tanksley SD, Zamir D (1995) Recombinant inbred lines for genetic mapping in tomato. Theor Appl Genet 90(3):542–548. https://doi.org/10.1007/BF00222001
Saxena RK, Kale SM, Kumar V et al (2017) Genotyping-by-sequencing of three mapping populations for identification of candidate genomic regions for resistance to sterility mosaic disease in pigeon pea. Sci Rep 7(1):1813. https://doi.org/10.1038/s41598-017-01535-4
Vikram P, Swamy BM, Dixit S, Ahmed HU, Cruz MTS, Singh AK, Kumar A (2011) qDTY 1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet 12(1):89. https://doi.org/10.1186/1471-2156-12-89
Rajurkar AB, Muthukumar C, Bharathi A, Thomas HB, Babu RC (2019) Saturation mapping of consistent QTLs for yield and days to flowering under drought using locally adapted landrace in rice (Oryza sativa L.). NJAS - Wageningen. J Life Sci 88:66–75. https://doi.org/10.1016/j.njas.2018.10.002
Marri PR, Sarla N, Reddy LV, Siddiq EA (2005) Identification and mapping of yield and yield related QTLs from an indian accession of Oryza rufipogon. BMC Genet 6(1):1–14. https://doi.org/10.1186/1471-2156-6-33
Cho YI, Jiang W, Chin JH, Piao Z, Cho YG, McCouch SR, Koh HJ (2007) Identification of QTLs associated with physiological nitrogen use efficiency in rice. Mol Cells 23(1):72–79
Baisakh N, Yabes J, Gutierrez A, Mangu V, Ma P, Famoso A, Pereira A (2020) Genetic mapping identifies consistent quantitative trait loci for yield traits of rice under greenhouse drought conditions. Genes 11(1):62. https://doi.org/10.3390/genes11010062
Luo LJ, Li ZK, Mei HW, Shu QY, Tabien R, Zhong DB, Ying CS, Stansel JW, Khush GS, Paterson AH (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice II grain yield components. Genetics 158(4):1755–1771. https://doi.org/10.1093/genetics/158.4.1737
Lafitte HR, Price AH, Courtois B (2004) Yield response to water deficit in an upland rice mapping population: associations among traits and genetic markers. Theor Appl Genet 109(6):1237–1246. https://doi.org/10.1007/s00122-004-1731-8
Fukai S, Pantuwan G, Jongdee B, Cooper M (1999) Screening for drought resistance in rainfed lowland rice. Field Crops Res 64(1–2):61–74. https://doi.org/10.1016/S0378-4290(99)00051-9
De Leon TB, Pruthi R, Jampala B, Borjas AH, Subudhi PK (2020) Genetic determinants for agronomic and yield-related traits localized on a GBS-SNP linkage map from a Japonica x Indica cross in rice. Plant Gene 24:100249. https://doi.org/10.1016/j.plgene.2020.100249
Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F (2003) Control of tillering in rice. Nature 422(6932):618–621. https://doi.org/10.1038/nature01518
Ali M, Pathan M, Zhang J, Bai G, Sarkarung S, Nguyen HJT (2000) Mapping QTLs for root traits in a recombinant inbred population from two indica ecotypes in rice. Theor Appl Genet 101:756–766. https://doi.org/10.1007/s001220051541
Chen Y, Li C, Zhang B, Yi J, Yang Y, Kong C, Lei C, Gong M (2019) The role of the late embryogenesis-abundant (lea) protein family in development and the abiotic stress response: a comprehensive expression analysis of potato (Solanum Tuberosum). Genes 10(2):148. https://doi.org/10.3390/genes10020148
Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115(1):35–46. https://doi.org/10.1007/s00122-007-0538-9
Wang J, Shi H, Zhou L, Peng C, Liu D, Zhou X, Wu W, Yin J, Qin H, Ma W, He M, Li W, Wang J, Li S, Chen X (2017) OsBSK1-2, an Orthologous of AtBSK1, is involved in Rice Immunity. Front Plant Sci 8908. https://doi.org/10.3389/fpls.2017.00908
Xiong Y, Gan L, Hu Y, Sun W, Zhou X, Song Z, Zhang X, Li Y, Yang Z, Xu W, Zhang J, He Y, Cai D (2019) OsMND1 regulates early meiosis and improves the seed set rate in polyploid rice. Plant Growth Regul 87(2):341–356. https://doi.org/10.1007/s10725-019-00476-4
Macovei A, Vaid N, Tula S, Tuteja N (2012) A new DEAD-box helicase ATP-binding protein (OsABP) from rice is responsive to abiotic stress. Plant Signal Behav 7(9):1138–1143. https://doi.org/10.4161/psb.21343
Yang SQ, Li WQ, Miao H, Gan PF, Qiao L, Chang YL, Shi CH, Chen KM (2016) REL2, a gene encoding an unknown function protein which contains DUF630 and DUF632 domains controls leaf rolling in rice. Rice 9(1):37. https://doi.org/10.1186/s12284-016-0105-6
Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G (2011) SnRK2 protein kinases–key regulators of plant response to abiotic stresses. OMICS 15(12):859–872. https://doi.org/10.1089/omi.2011.0091
Gho YS, Park SA, Kim SR, Chandran AKN, An G, Jung KH (2017) Comparative expression analysis of rice and arabidopsis peroxiredoxin genes suggests conserved or diversified roles between the two species and leads to the identification of tandemly duplicated rice peroxiredoxin genes differentially expressed in seeds. Rice 10(1):30. https://doi.org/10.1186/s12284-017-0170-5
Acknowledgements
The authors gratefully acknowledge the Department of Biotechnology, and DBT-NECAB, Assam Agricultural University, Jorhat, Assam for providing the financial support for conducting this work. Rahul Kaldate gratefully acknowledges Indian Council of Agricultural Research, New Delhi for providing Senior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaldate, R., Verma, R.K., Chetia, S.K. et al. Mapping of QTLs associated with yield and related traits under reproductive stage drought stress in rice using SNP linkage map. Mol Biol Rep 50, 6349–6359 (2023). https://doi.org/10.1007/s11033-023-08550-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08550-x