Abstract
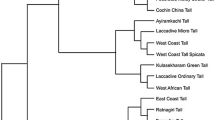
A total of 12,080 sequences, including 5,851 transcriptome contigs developed at NBRI and 5,002 singlets and 1,227 contigs assembled from 13,201 expressed sequence tags (ESTs) of Jatropha curcas from National Center for Biotechnology Information database were used to search for simple sequence repeats (SSRs). Seven hundred and two sequences containing 786 SSRs with 93.4% simple and 7.6% compound repeat motifs were identified. Dinucleotide repeats (DNRs) were most abundant, followed by trinucleotide and tetranucleotide repeats. AG/CT was the most common motif (50.0%) followed by AT/AT (38.8%) and AC/GT (10.0%) among the DNRs. Four hundred and six primer pairs were designed out of the 702 SSR-containing sequences. Fifty randomly selected EST-SSR markers were amplified in 25 accessions collected from different geographical regions of India. Twenty-one SSR markers were polymorphic and with allele variation from two to four. Polymorphic information content value ranged between 0.04 and 0.61 with an average of 0.25 ± 0.16, indicating low to moderate level of informativeness within these EST-SSRs. The polymorphic markers showed 57.0% to 95.6% transferability among five species of Jatropha and 47.0% transferability across genera in Ricinus communis. Fifty-one alleles detected by the 21 polymorphic EST-SSRs were used to determine genetic relationships among 25 J. curcas accessions. Genetic similarity coefficient ranged from 0.44 to 0.94. The 25 accessions got grouped to three main clusters, comprising 10, 11, and four accessions. This is the first report of development of EST-SSRs in J. curcas and will be valuable resource for future genetical studies, like construction of linkage maps, diversity analysis, quantitative trait locus/association mapping, and molecular breeding of J. curcas.



Similar content being viewed by others
References
Aggarwal RK, Hendre PS, Varshney RK, Bhat PR, Krishnakumar V, Singh L (2007) Identification, characterization and utilization of EST-derived genic microsatellite markers for genome analyses of coffee and related species. Theor Appl Genet 114:359–372
Anderson JR, Lubberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Basha SD, Sujatha EM (2007) Inter and intra-population variability of Jatropha curcas (L.) characterized by RAPD and ISSR markers and development of population specific SCAR markers. Euph 156:375–386
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Cardle L, Ramsay L, Milbourne D, Macaulay M, Marshall D, Waugh R (2000) Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genet 156:847–854
Carvalho CR, Clarindo WR, Prac MM, FS Araujo, Carels N (2008) Genome size, base composition and karyotype of Jatropha curcas L.: an important biofuel plant. Plant Sci 174:613–617
Chabane M, Ablett GA, Cordeiro GM, Valkoun J, Henry RJ (2005) EST versus genomic derived microsatellite markers for genotyping wild and cultivated barley. Genet Resour Crop Evol 52:903–909
Chapman MA, Hvala J, Strever J, Matvienko M, Kozik A, Michelmore RW, Tang S, Knapp SJ (2009) Development, polymorphism, and cross-taxon utility of EST–SSR markers from safflower (Carthamus tinctorius L.). Theor Appl Genet 120:85–91
Chen CX, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from citrus ESTs. Theor Appl Genet 112:1248–1257
Cho YG, Ishii T, Temmykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and Genebank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euph 142:169–196
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellites markers from sugarcane (Saccharum spp.) ESTs cross transferable to Erianthus and Sorghum. Plant Sci 160:1115–1123
Feng SP, Li WG, Huang HS, Wang JY, Wu YT (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23:85–97
Field D, Wills C (1998) Abundant microsatellite polymorphisms in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc Nat Acad Sci U S A 95:1647–1652
Foidl N, Elder P (1997) Agro-industrial exploitation of Jatropha curcas. In: Gubitz GM, Mittelbach M, Trabi M (eds) Biofuel and industrial products from Jatropha curcas. Dvb, Graz
Ganesh RS, Parthiban KT, Senthil Kumar R, Thiruvengadam V, Paramathma VM (2008) Genetic diversity among Jatropha species as revealed by RAPD markers. Genet Resour Crop Evol 55:803–809
Gao LF, Tang J, Li H, Jia J (2003) Analysis of microsatellites in major crops assessed by computational and experimental approaches. Mol Breed 113:163–185
Gao LZ, Zhang CH, Jia JZ (2005) Cross-species transferability of rice microsatellites in its wild relatives and the potential for conservation genetic studies. Genet Resour Crop Evol 52:931–940
Ginwal HS, Phartyal SS, Rawat PS, Srivastava RL (2005) Seed source variation in morphology, germination and seedling growth of Jatropha curcas L. in central India. Silvae Genet 54(2):76–80
Girach RD, Ahmad A, Baghrendah M (1995) Jatropha curcas L.: a potential source of herbal drug in dental complaints. Hamdard Med 38:94–100
Gupta PK, Roy JK (2002) Molecular markers in crop improvement: present status and future needs in India. Plant Cell Tissue Organ Cult 70:229–234
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Gen Genomics 270:315–323
Han ZG, Wang CB, Song XL, Guo WZ, Gou JY, Li CH, Chen XY, Zhang TZ (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST-SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Henning R (1998) Use of Jatropha curcas—household perspective and its contribution to rural employment creation. In: Proceedings of the Regional Workshop on the “Potential of Jatropha curcas in Rural Development and Environmental Protection,” May 13–15, Harare, Zimbabwe
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877
Juan L, Fang Y, Lin T, Fang C (2003) Antitumor effects of curcin from seeds of Jatropha curcas. Acta Pharmacol Sin 24:241–246
Jung S, Albert A, Jesudurai C, Tomkins J, Main D (2005) Frequency, type, distribution and annotation of simple sequence repeats in Rosaceae ESTs. Funct Integr Genomics 5:136–143
Kantety RV, Rota ML, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barely, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kaushik N, Kumar K, Kumar S, Kaushik N, Roy S (2007) Genetic variability and divergence studies in seed traits and oil content of Jatropha (Jatropha curcas L.) accessions. Biomass Bioenergy 31:497–502
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998
Lakshmanan P, Mohan S, Jeyarajan R (1990) Antifungal properties of some plant extracts against Thanatephorus cucumeris, the causal agent of collar rot disease of Phaseolus aureus. Madras Agric J 77:1–4
Liang X, Chen X, Hong Y, Liu H, Zhou G, Li S, Guo B (2009) Utility of EST-derived SSR in cultivated peanut (Arachis hypogaea L.) and Arachis wild species. BMC Plant Biol 9:35
Liu K, Muse SV (2005) Power marker: integrated analysis environment for genetic marker data. Bioinformatics 21:2128–2129
Luro FL, Costantino G, Terol J, Argout X, Allario T, Wincker P, Talon M, Ollitrault P, Morillon R (2008) Transferability of the EST–SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9:287
Marroquin EA, Bainco JA, Granados S, Caceres A, Morales C (1997) Clinical trial of Jatropha curcas sapin treatment of common warts. Fitoterapia 68:160–162
Nicot N, Chiquet V, Gandon B, Amilhat L, Legeai F, Leroy P, Bernard M, Sourdille P (2004) Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs). Theor Appl Genet 109:800–805
Openshaw K (2000) A review of Jatropha curcas: an oil plant of unfulfilled promise. Biomass Bioenergy 19:1–15
Pan L, Xia Q, Quan Z, Liu H, Ke W, Ding Y (2010) Development of novel EST-SSRs from sacred lotus (Nelumbo nucifera Gaertn) and their utilization for the genetic diversity analysis of N. nucifera. J Heredity 101:71–82
Parthiban KT, Senthil Kumar R, Thiyagarajan P, Subbulakshmi V, Vennila S, Govinda Rao M (2009) Hybrid progenies in Jatropha—a new development. Curr Sci 96:815–823
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Enfield, Montpellier, pp 43–76
Powell W, Machray G, Provan J (1996a) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996b) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Raji AAJ, Anderson JV, Kolade OA, Ugwu CD, Dixon AGO, Ingelbrecht IL (2009) Gene-based microsatellites for cassava (Manihot esculenta Crantz): prevalence, polymorphisms, and cross-taxa utility. BMC Plant Biol 9:118
Ranade SA, Srivastava AP, Rana TS, Srivastava J, Tuli R (2008) Easy assessment of diversity in Jatropha curcas L. plants using two single-primer amplification reaction (SPAR) methods. Biomass Bioenergy 32:533–540
Rug M, Ruppel A (2000) Toxic activities of the plant Jatropha curcas against intermediate snail hosts and larvae of schistosomes. Trop Med Int Hlth 5:423–430
Rungis D, Berube Y, Zhang J, Ralph S, Ritland CE, Ellis BE, Douglas C, Bohlmann J, Ritland K (2004) Robust simple sequence repeats markers for spruce (Picea spp.) from expressed sequence tags. Theor Appl Genet 109:1283–1294
Schuelke M (2000) An economic method for the fluorescent labelling of PCR fragments. Nat Biotechnol 18:233–234
Scott KD, Eggler P, Seaton G, Rossetto EM, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Sharma RK, Bhardwaj P, Negi R, Mohapatra T, Ahuja PS (2009) Identification, characterization and utilization of unigene derived microsatellite markers in tea (Camellia sinensis L.). BMC Plant Biol 9:53
Subramanian KA, Singal SK, Saxena M, Singhal S (2005) Utilization of liquid biofuels in automotive diesel engines: an Indian perspective. Biomass Bioenergy 29:65–72
Sudheer DVNP, Pandya N, Reddy MP, Radhakrishnan T (2008) Comparative study of interspecific genetic divergence and phylogenic analysis of genus Jatropha by RAPD and AFLP. Mol Biol Rep 36:901–907
Sudheer DVNP, Sinha R, Kothari P, Reddy MP (2009) Isolation of novel microsatellites from Jatropha curcas L. and their cross-species amplification. Mol Eco Resour 9:431–433
Sujatha M, Prabakaran AJ (1997) Characterization and utilization of Indian Jatrophas. Indian J Plant Genet Resour 10:123–128
Sujatha M, Prabakaran AJ (2003) New ornamental Jatropha hybrids through interspecific hybridization. Genet Resour Crop Evol 50:75–82
Sun QB, Li LF, Li Y, Wu GJ, Ge XJ (2008) SSR and AFLP markers reveal low genetic diversity in the biofuel plant Jatropha curcas in China. Crop Sci 48:1865–1871
Tatikonda L, Wani SP, Kannan S, Beerelli N, Sreedevi TK, Hoisington DA, Devi P, Varshney RK (2009) AFLP-based molecular characterization of an elite germplasm collection of Jatropha curcas L., a biofuel plan. Plant Sci 176:505–513
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST database for the development and characterization of gen-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Toth G, Gaspari Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981
Varshney RK, Graner A, Sorrells ME (2005a) Genic microsatellite markers: features and applications. Trends Biotechnol 23:48–55
Varshney RK, Sigmund R, Borner A, Korzun V, Stein N, Sorrells ME (2005b) Inter-specific transferability and comparative mapping of barley EST-SSR markers in wheat, rye and rice. Plant Sci 168:195–202
Zhang LY, Bernard M, Leroy P, Feuillet C (2005) High transferability of bread wheat EST-derived SSRs to other cereals. Theor Appl Genet 111:677–687
Acknowledgements
The authors thank the Council of Scientific and Industrial Research/NMITLI program under which the different accessions were collected. AR thanks CSIR for the award of Senior Research Fellowship. RT thanks DST for JC Fellowship. Useful suggestions by Dr. JK Roy, during manuscript preparation, are also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Dandekar
Rights and permissions
About this article
Cite this article
Kumar Yadav, H., Ranjan, A., Asif, M.H. et al. EST-derived SSR markers in Jatropha curcas L.: development, characterization, polymorphism, and transferability across the species/genera. Tree Genetics & Genomes 7, 207–219 (2011). https://doi.org/10.1007/s11295-010-0326-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-010-0326-6