Abstract
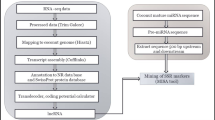
Genetic improvement in coconut relies on exploiting the vast existing diversity among coconut accessions. Robust molecular markers are a pre-requisite for efficient characterization of genetic diversity. Microsatellites or simple sequence repeats (SSRs), mined from expressed sequence tags (ESTs), constitute an important resource for analysis of genetic diversity as they are abundant, polymorphic and represent function regions of the genome. We have identified a total of 318,528 putative EST-SSRs from 130,942 unigenes utilizing a leaf transcriptome dataset of coconut. Among the EST-SSRs, dinucleotide repeats were abundant (219,912; 69.04%) followed by trinucleotide (70,722; 22.2%) and tetra-nucleotide repeats (6281; 1.9%). Among the dinucleotide repeat motifs, the dominant repeat was AG/CT (35.87%), followed by AT/AT (18.59%), while the dominant trinucleotide repeat was AAG/CTT (4.59%). One hundred and twenty EST-SSR primer pairs were designed and utilized to amplify six DNA samples of coconut accessions. Fifty primers (41.7%) produced reproducible polymorphic fragments of expected sizes, from which a total of 10 primers were selected for the diversity assessment in 186 palms of 50 coconut accessions, comprising of 25 each of tall and dwarf accessions. A total of 137 alleles were detected with an average of 13.7 alleles per SSR locus. The number of alleles observed at each locus in the data set ranged from 7 to 22. All the loci showed 100% polymorphism with respect to the samples screened. The average observed heterozygosity was 0.46. The PIC values ranged from 0.79 (CnKGDEST129 and CnKGDEST100) to 0.91 (CnKGDEST117 and CnKGDEST122) with a mean value of 0.85, indicating the capacity of the EST-SSR markers to detect high levels of polymorphism. The cluster analysis revealed that accessions were generally clustered based on their relative similarity and irrespective of their geographic origins. The present study demonstrates the usefulness of transcriptome sequencing as a rapid and cost-effective methodology for the development of molecular markers. The EST-SSR markers generated through this study constitute useful and reliable tools for assessment of genetic diversity and marker-assisted selection in coconut.


Similar content being viewed by others
References
Rajesh MK, Sabana AA, Rachana KE, Rahman S, Jerard BA, Karun A (2015) Genetic relationship and diversity among coconut (Cocos nucifera L.) accessions revealed through SCoT analysis. 3 Biotech 5(6):999-1006
Arunachalam V, Rajesh MK (2017) Coconut genetic diversity, conservation and utilization. In: Ahuja MR, Jain SM (eds) Biodiversity and conservation of woody plants. Springer, Cham, pp 3–36
Nair RV, Jerard BA, Thomas RJ (2016) Coconut breeding in India. In: Al-Khayari JM, Jain SM (eds) Advances in plant breeding strategies: agronomic, abiotic and biotic stress traits. Springer, Cham, pp 257–279
Lebrun P, N’Cho YP, Seguin M, Grivet L, Baudouin L (1998) Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers. Euphytica 101(1):103–108
Qiu L, Yang C, Tian B, Yang JB, Liu A (2010) Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.). BMC Plant Biol 10(278):1–10
Rao VR, Hodgkin T, Bourdeix R (2005) Locating coconut genetic diversity. In: Batugal P, RamananthaRao V, Oliver J (eds) Coconut genetic resources. International Plant Genetic Resources Institute, Regional Office for Asia, the Pacific and Oceania (IPGRI-APO), Serdang, pp 13–31
Arunachalam V, Jerard BA, Damodaran V, Ratnambal MJ, Kumaran PM (2005) Phenotypic diversity of foliar traits in coconut germplasm. Genet Resour Crop Ev 52(8):1031–1037
Sugimura Y, Itano M, Salud CD, Otsuji K, Yamaguchi H (1997) Biometric analysis on diversity of coconut palm: cultivar classification by botanical and agronomical traits. Euphytica 98:29–35
Geethalakshmi P, Parthasarathy VA, Niral V (2005) Genetic diversity among coconut (Cocos nucifera L.) genotypes using isozymes. Asian J Plant Sci 4:678–683
Kumar P, Gupta VK, Misra AK, Modi DR, Pandey BK (2009) Potential of molecular markers in plant biotechnology. Plant Omics 2(4):141–162
Teulat B, Aldam C, Trehin R, Lebrun P, Barker JHA, Arnold GM, Karp A, Baudouin L, Rognon F (2000) An analysis of genetic diversity in coconut (Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. TheorAppl Genet 100(5):764–771
Merrow AW, Wisser RJ, Brown SJ, Kuhn DN, Schnell RJ, Broschat TK (2003) Analysis of genetic diversity and population structure within Florida coconut (Cocos nucifera L.) germplasm using microsatellite DNA, with special emphasis on the Fiji Dwarf cultivar. Theor Appl Genet 106(4):715–726
Perera L, Russell JR, Provan J, Powell W (2000) Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.). Genome 43(1):15–21
Perera L, Russell JR, Provan J, Powell W (2001) Levels and distribution of genetic diversity of coconut (Cocos nucifera L. var Typica form typica) from Sri Lanka assessed by microsatellite markers. Euphytica 122(2):381–389
Rajesh MK, Karun A, Parthasarathy VA (2019) Coconut biotechnology. In: Nampoothiri KUK, Krishnakumar V, Thampan PK, Nair MA (eds) The coconut palm (Cocos nucifera L.)-research and development perspectives. Springer, Singapore, pp 191–226
Rajesh MK, Arunachalam V, Nagarajan P, Lebrun P, Samsudeen K, Thamban C (2008) Genetic survey of 10 Indian coconut landraces by simple sequence repeats (SSRs). Sci Hortic 118(4):282–287
Rivera R, Edwards KJ, Barker JHA, Arnold GM, Ayad G, Hodgkin T, Karp A (1999) Isolation and characterization of polymorphic microsatellites in Cocos nucifera L. Genome 42(4):668–675
Xia W, Xiao Y, Liu Z, Luo Y, Mason AS, Fan H, Yang Y, Zhao S, Peng M (2014) Development of gene-based simple sequence repeat markers for association analysis in Cocos nucifera. Mol Breed 34(2):525–535
Xiao Y, Luo Y, Yang Y, Fan H, Xia W, Mason AS, Zhao S, Sager R, Qiao F (2013) Development of microsatellite markers in Cocos nucifera and their application in evaluating the level of genetic diversity of Cocos nucifera. Plant Omics 6(3):193–200
Senan S, Kizhakayil D, Sasikumar B, Sheeja TE (2014) Methods for development of microsatellite markers: an overview. Not Sci Biol 6(1):1–13
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23(1):48–55
Casu R, Dimmock C, Thomas M, Bower N, Knight D, Grof C (2001) Genetic and expression profiling in sugarcane. Proc Int Soc Sugarcane Technol 24:626–627
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Gene 12(7):499–510
Rajesh MK, Rachana KE, Kulkarni K, Sahu BB, Thomas RJ, Karun A (2018) Comparative transcriptome profiling of healthy and diseased Chowghat Green Dwarf coconut palms from root (wilt) disease hot spots. Eur J Plant Pathol 151(1):173–193
Rajesh MK, Jerard BA, Preethi P, Thomas RJ, Fayas TP, Rachana KE, Karun A (2013) Development of a RAPD-derived SCAR marker associated with tall-type palm trait in coconut. Sci Hortic 150:312–316
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19(2):153–170
Lewis PO, Zaykin D (2001) Genetic Data Analysis: computer program for the analysis of allelic data. Version 1.0.
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JP, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9(1):6
Feng SP, Li WG, Huang HS, Wang JY, Wu YT (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23(1):85–97
Ting NC, Noorhariza MZ, Rozana R, Low ET, Ithnin M, Cheah SC, Tan SG (2010) SSR mining in oil palm EST database: application in oil palm germplasm diversity studies. J Genet 89:135–145
Varshney RK, Thiel T, Stein N, Langridge P, Graner A (2002) In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell Mol Biol Lett 7(2):537–546
Wang YW, Samuels TD, Wu YQ (2011) Development of 1,030 genomic SSR markers in switch grass. Theor Appl Genet 122(4):677–686
Li YC, Korol AB, Fahima T, Nevo E (2004) Microsatellites within genes: structure, function, and evolution. Mol Biol Evol 21:991–1007
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10(1):72–80
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48(5–6):501–510
Gong YM, Xu SC, Mao WH, Hu QZ, Zhang GW, Ding J, Li ZY (2010) Generation and characterization of 11 novel EST derived microsatellites from Vicia faba (Fabaceae). Am J Bot 97(7):e69–e71
Kuroda Y, Tomooka N, Kaga A, Wanigadeva SMSW, Vaughan DA (2009) Genetic diversity of wild soybean and Japanese cultivated soybeans based on microsatellite (SSR) analysis and the selection of a core collection. Genet Resour Crop Ev 56(8):1045–1055
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177(3):309–334
Wang Z, Yu G, Shi B, Wang X, Qiang H, Gao H (2014) Development and characterization of simple sequence repeat (SSR) markers based on RNA-sequencing of Medicago sativa and in silico mapping onto the M. truncatula genome. PLoS ONE 9(3):e92029
Rajesh MK, Nagarajan P, Jerard BA, Arunachalam V, Dhanapal R (2008) Microsatellite variability of coconut accessions from Andaman and Nicobar Islands. Curr Sci 94(12):1627–1631
Zizumbo-Villarreal D, Colunga-GarcíaMarín P (2001) Morpho-physiological variation and phenotypic plasticity in Mexican populations of coconut (Cocos nucifera L.). Genet Resour Crop Evol 48(6):547–554
Devakumar K, Neema BTS, Uma M, Naganeeswaran S, Niral V, Jerard BA (2016) Analysis of genetic diversity among Indian Ocean coconut accessions through microsatellite markers. Indian J Hort 73(1):13–18
Niral V, Samsudeen K, Sudha R, Ranjini TN (2019) Genetic resource management and improved varieties of coconut. Ind Cocon J 28:10–14
Samsudeen K, Jacob PM, Rajesh MK, Jerard BA, Kumaran PM (2006) Origin and evolution and evolution of Laccadive Micro Tall, a coconut cultivar from Lakshadweep Islands of India. J Plant Crops 34(3):220–225
Harries HC (1978) The evolution, dissemination and classification of Cocos nucifera L. Bot Rev 44:205–315
Jerard BA, Damodaran V, Niral V, Samsudeen K, Rajesh MK, Sankaran M (2013) Conservation and utilization of soft endosperm coconut accession from Andaman Islands. J Plant Crops 41(1):14–21
Funding
This research was funded by Indian Council of Agricultural Research (ICAR-CPCRI Institute Project Code No. 1000761030).
Author information
Authors and Affiliations
Contributions
MKR designed the study; BAJ and VN provided the samples; PP, SR and SN performed the experiments; PP, AAS, KPG and MKR analyzed the data; PP wrote the paper; MKR, VN and BAJ checked the paper. The decision to submit the manuscript for publication was made by all the authors. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
None of the authors have any financial or non-financial conflict of interest.
Research involving human and/or animals rights
This research did not involve Human Participants and/or Animals.
Informed consent
Not applicable as this research did not involve human participants or clinical trials.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Preethi, P., Rahman, S., Naganeeswaran, S. et al. Development of EST-SSR markers for genetic diversity analysis in coconut (Cocos nucifera L.). Mol Biol Rep 47, 9385–9397 (2020). https://doi.org/10.1007/s11033-020-05981-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05981-8