Abstract
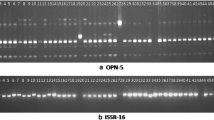
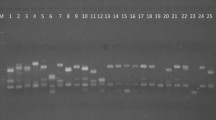
Genus Jatropha with 172 species having significant economic importance belongs to the family Euphorbiaceae. There are no reports on molecular characterization and phylogenetic relationship among the species of Jatropha. Hence, the present study was undertaken to assess the extent of genetic variability that exist and also to establish phylogenetic relationship among Jatropha curcas, J. glandulifera, J. gossypifolia, J. integerrima, J. multifida, J. podagrica and J. tanjorensis using RAPD and AFLP. The percentage of loci that are polymorphic among the species studied was found to be 97.74% by RAPD and 97.25% by AFLP. The mean percentage of polymorphism (PP) was found to be 68.48 by RAPD and 71.33 by AFLP. The phylogram generated with RAPD and AFLP data showed maximum similarity. With the generated data maximum relatedness was found between J. curcas and J. integerrima this may be the reason for the success of inter hybrid crosses between these two species. Neither RAPD nor AFLP data generated in this study supports the view of J. tanjorensis, a natural interspecific hybrid between J. curcas and J. gossypifolia. The present study concludes that both RAPD and AFLP techniques are comparable in divergence studies of Jatropha species. The markers generated by RAPD and AFLP can be employed efficiently for interspecific hybrids identification, marker assisted selection and genetic resource management.




Similar content being viewed by others
References
Fairless D (2007) Biofuel: the little shrub that could - maybe. Nature 449:652–655
Sunita K, Kochar VK, Singh SP, Katiyar RS, Pushpangadan P (2005) Differential rooting and sprouting behaviour of two Jatropha species and associated physiological and biochemical changes. Curr Sci 89:936–939
Olapeju OA, Kayode A, Olusegun E, James BG (2007) Antibacterial diterpenoids from Jatropha podagrica Hook. Phytochemistry 68:2420–2425
Prabakaran AJ, Sujatha M (1999) Jatropha tanjorensis Ellis & Saroja, a natural interspecific hybrid occurring in Tamil Nadu, India. Gene Res Crop Evol 46:213–218
Yotam L, Yaniv S, Haim B, Menachem S, Emile H (2000) Rare Jatropha multifida intoxication in two children. J Emerg Med 19:173–175
Taofeeq O, Ganiyu OA, Tesleem AO, Godwin OA, Mutiyat AO (2005) Mechanism of action of Jatropha gossypifolia stem latex as a haemostatic agent. Eur J Gen Med 2(4):140–143
Ramchandani M, Jolly CI (1988) Pharmacognostical and phytochemical studies on Phyllanthus fraternus webster and Jatropha glandulifera ROXB. Indian J Pharm Sci 50:276–277
Banerji R, Chowdhury AR, Misra G, Sudarsanam G, Verma SC, Srivastava GS (1985) J. curcas seed oil for energy. Biomass 8:277–282
Gubitz GM, Mittelbach M, Trabi M (1999) Exploitation of the tropical oil seed plant Jatropha curcas L. Bioresour Technol 67:73–82
Mandpe S, Kadlaskar S, Degen W, Keppeler S (2005) On road testing of advanced common Rail diesel vehicles with biodiesel from the Jatropha Curcas plant, Society of Automotive Engineers. Inc 26:356–364
Martin G, Mayeux A (1984) Réflexions sur les cultures oléagineuses énergétiques. II. - Le Pourghère (Jatropha curcas L.): UN carburant possible. Oléagineux 39:283–287
Staubmann R, Ncube I, Gubitz GM, Steiner W, Read JS (1999) Esterase and lipase activity in Jatropha curcas L. J Biotechnol 75:117–126
Takeda Y (1982) Development study on Jatropha curcas (Sabudum) oil as a substitute for diesel engine oil in Thailand. J Agric Assoc China 120:1–8
Ginwal HS, Rawat PS, Srivastava RL (2004) Seed source variation in growth performance and oil yield of Jatropha curcas Linn. in central India. Silve Senetica 53:186–192
Doyle JJ, Doyle JL (1990) Isolation of fresh DNA from fresh tissue. Focus 12:3–15
Williams JG, Kubelik AR, Livak J, Rafalski J, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Vos P, Hogers R, Bleeker M, Reijans M, de Lee TV, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Jaccard P (1908) Nouvelles recherchesur la distribution florale. Bull Soc Vaud Sci Natl 44:223–270.
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Francisco-Ortega J, Crawford DJ, Santos-Guerra A, Cravalho JA (1996) Isozyme differentiation in the endemic genus Argyranthemum (Asteraceae, Anthemideae) in the Macaronesian islands. Plant Syst Evol 202:137–152
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In Brown HD et al (eds) Plant population genetics, breeding and genetic resources. Sinaue Sunderland, MA, USA, pp 43–63
Crawford DJ, Haines DW, Cosmer MB, Wiens D, Lopez P (1994) Lactoris fernandeziana on the Juan Fernandez Islands allozyme uniformity and field obsevarvations. Conserv Biol 8:277–280
Essilman EJ, Crawford DJ, Brauner S, Stuessy TF, Anderson GJ, Silva OM (1997) RAPD marker diversity within and divergence among species of Dendroseris (Asteraceae: Lactuceae). Am J Bot 4:591–596
Francisco-Ortega J, Crawford DJ, Santos-Guerra A, Cravalho JA (1996) Isozyme differentiation in the endemic genus Argyranthemum (Asteraceae, Anthemideae) in the Macaronesian islands. Plant Syst Evol 202:137–152
Lesica P, Leary RF, Allendort FR, Bilderbecl DE (1998) Lac of genetic diversity within and among populations of an endangered plant, Hawellia aquatilis. Conserv Biol 2:275–282
Lowrey TK, Crawroford DJ (1985) Allozyme divergence and evolution in Tertamolopium (Compositae; Astereae) on the Hawaiian Island. Syst Bot 10:64–72
Soltis PS, Soltis DE, Tucker TL, Lang FA (1992) Allozyme variability is absent in the narrow endemic Bensoniella oregona (Saifragacear). Conserv Biol 6:131–134
Adams RP, Demeke T (1993) Systematic relationships in Juniperus based on random amplified polymorphic DNAs (RAPDs). Taxon 42:553–571
Campos LP, Raelson JV, Grant WF (1994) Genome relation ships among lotus species based on random amplified polymorphic DNA (RADP). Theor Appl Genet 88:417–422
Catalan P, Shi Y, Armstrong L, Draper J, Stace CA (1995) Molecular phylogeny of the grass genus Brachypodium P. beauv. based on RFLP and RAPD analysis. Bot J Linn Soc 117:263–280
Essilman EJ, Crawford DJ, Brauner S, Stuessy TF, Anderson GJ, Silva OM (1997) RAPD marker diversity within and divergence among species of Dendroseris (Asteraceae: Lactuceae). Am J Bot 4:591–596
Ranade SA, Kumar A, Goswami M, Farooqui N, Sane ΡV (1997) Genome analysis of Amaranths: determination of inter and intra-species variations. J Biosci 22:457–464
Spooner DM, Ugarte L, Scroch PW (1997) Species boundaries and interrelationships of two closely related sympatric diploid wild potato species, Solanum astleyl and Solanum bollviense, based on RAPDs. Theor Appl Genet 96:764–771
Brucci G, Menozzi P (1993) Segregation analysis of random amplified polymorphic DNA (RAPD) markers in Picea abies Karst. Mol Ecol 2:227–232
Brunell MS, Whitkus R (1997) RAPD marker veration in Eriastrum densifolium (polemoniaceae), implications for sub specific delimitation and conservation. Syst Bot 22:543–553
Volis S, Yakubov B, Shulagina I, Ward D, Zur V, Mendlinger S (2001) Tests for adaptive RAPD variation in population genetic structure of wild barley, Hordeum spontaneum Koch. Biol J Linn Soc 74:289–303
Brunell MS, Whitkus R (1997) RAPD marker veration in Eriastrum densifolium (Polemoniaceae), implications for sub specific delimitation and conservation. Syst Bot 22:543–553
Hadrys H, Balick M, Schierwater B (1992) Applications of random amplified polymorphic DNA(RAPD) in molecular ecology. Mol Ecol 1:55–263
Basha SD, Sujatha M (2007) Inter and intra-population variability of Jatropha curcas (L.) characterized by RAPD and ISSR markers and development of population-specific SCAR markers. Euphytica 156:375–386
Ganesh RS, Parthiban KT, Senthil KR, Thiruvengadam V, Paramathma M (2008) Genetic diversity among Jatropha species as reveled by RAPD markers. Genet Resour Crop Evol. doi:10.1007/s10722-007-9285-7
Sujatha M, Prabakaran AJ (2003) New ornamental Jatropha hybrids through interspecific hybridization. Genet Resour Crop Evol 50:75–82
Sujatha M, Makkar HPS, Becker K (2005) Shoot bud proliferation from axillary nodes and leaf sections of non-toxic Jatropha curcas L. Plant Growth Regul 47:83–90
Soontornchinaksang P, Jenjittikul T (2003) Karyology of Jatropha (Euphorbiaceae). Thai For Bull (Bot) 31:105–112
Rupert EA, Dehgan B, Webster GL (1970) Experimental studies of relationships in the genus Jatropha. I. J. curcas X J. integerrima. Bull Torrey Bot Club 99:321–325
Bussel JD, Michelle W, Jennifer AC (2005) Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspect Plant Ecol Evol Syst 7:3–26
Acknowledgements
We are grateful to Dr. P. K. Ghosh, Director CSMCRI for encouragement and Dimyler Chryslar, Germany and CSIR New Delhi for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sudheer Pamidiamarri, D.V.N., Pandya, N., Reddy, M.P. et al. Comparative study of interspecific genetic divergence and phylogenic analysis of genus Jatropha by RAPD and AFLP. Mol Biol Rep 36, 901–907 (2009). https://doi.org/10.1007/s11033-008-9261-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-008-9261-0